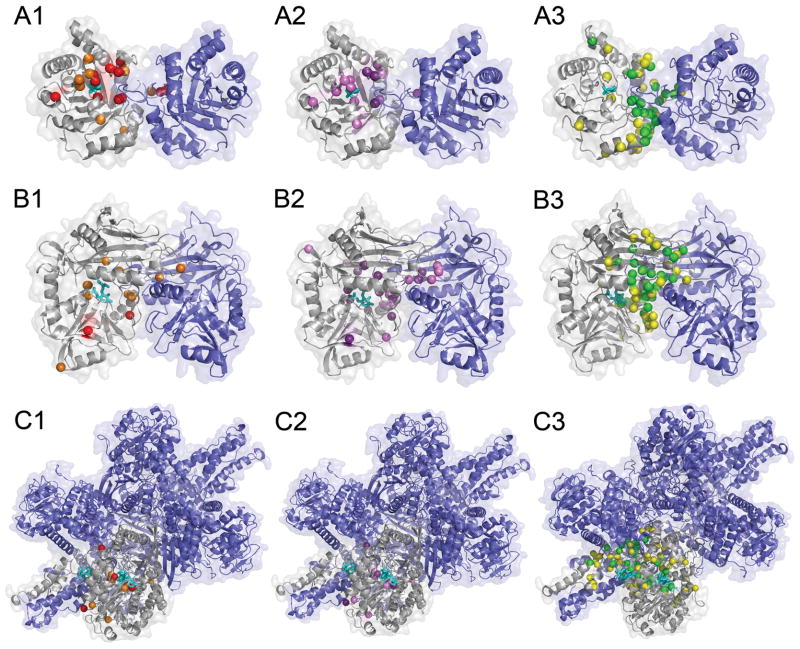
Figure 8.
Comparison of residues selected by naive sequence conservation, ConSurf, and BindML. A, triosephosphate isomerase homo-dimer (7TIM); B, amino acid transferase homo-dimer (1KT8); C, glutamate dehydrogenase (1HWZ). Ligand molecules binding to these proteins are shown in cyan. For the three proteins, residues which are assigned with a significantly high score by the three methods are shown. A1, B1, C1, residues with high conservation; A2, B2, C2, residues with a high score by ConSurf; A3, B3, C3, residues identified by BindML. For sequence conservation, residues which are conserved in more than 90% (70%) of sequences are shown in red (orange) (A1, B1, C1). As for ConSurf, residues detected with a Z-score of −1.3 (−1) or lower is shown in purple (violet) (A2, B2, C2). Residues identified with a Z-score of −1 (−0.5) or lower by BindML are shown in gree (yellow) (A3, B3, C3).