Figure 2. Construction of molecular networks.
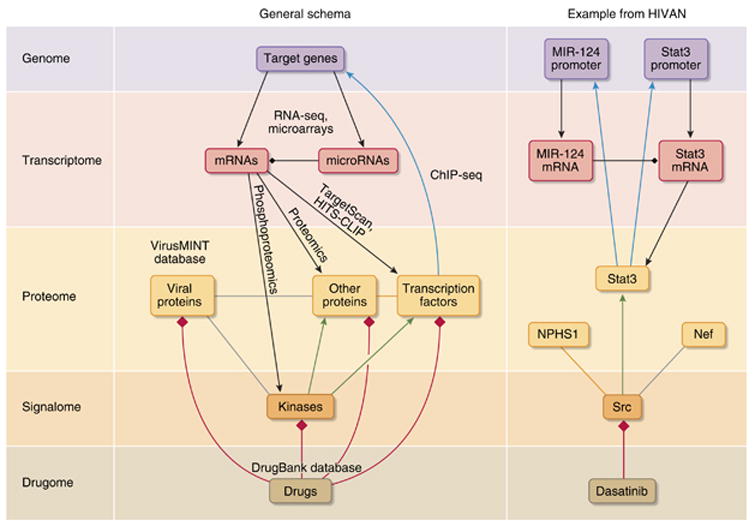
Networks that integrate binary interactions from heterogeneous sources, such as microRNAs (miRNAs) targeting mRNAs (from databases such as TargetScan or through experiments such as Argonaute HITS-CLIP124 (black diamond heads), protein/DNA interactions based on ChIP followed by high-throughput sequencing (ChIP-seq) analyses (cyan arrows), protein–protein interactions (orange links)125, virus/host protein interactions from databases such as VirusMINT75 (gray lines), kinase–substrate interactions126 (green arrows), and drug–target interactions76 (red diamond heads) from databases such as the DrugBank127, can be used to build networks made of different types of nodes and links. An example of such network for key components that have a role in human immunodeficiency virus-associated nephropathy (HIVAN) is given on the right. Dasatinib is an Src inhibitor used for the treatment of oncologic disorder, but could be tested for its therapeutic potential in the treatment of HIV-associated nephropathy. Arrowheads indicate activations and diamond heads denote inhibition. NPHS1, nephrosis 1 homolog.
