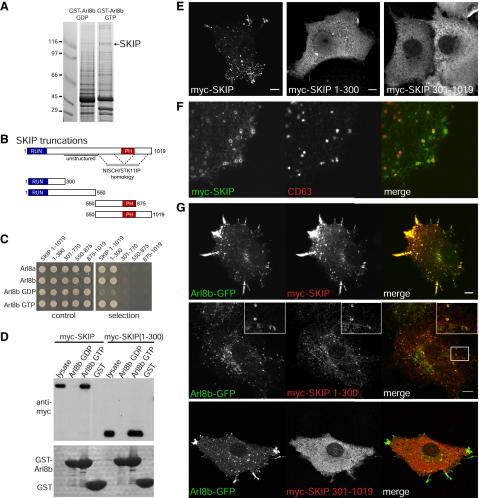
Figure 1.
Arl8 Binds to the RUN Domain of SKIP, and the Two Proteins Colocalize on Lysosomes
(A) Coomassie-stained protein gel of eluates from large-scale affinity chromatography of HeLa cytosol using GST fusions to Arl8b-T34N (GDP) or Arl8b-Q75L (GTP). Each lane represents 1.6 × 108 cells. Bands associated with Arl8b-Q75L were excised and mass spectrometry of tryptic peptides identified seven sequences corresponding to SKIP. Other bands examined were heat shock proteins or abundant cytosolic enzymes.
(B) Schematic of the organization of SKIP indicating the RUN and PH domains, and the region predicted to be unstructured. The regions flanking the PH domain are distantly related to a region of unknown function found at the C-termini of nischarin and STK11IP (serine/threonine kinase 11 interacting protein) (Lim and Hong, 2004).
(C) Yeast two-hybrid interactions between Arl8a or Arl8b, or the latter with T34N (GDP) or Q75L (GTP) mutations, and SKIP or the indicated truncations.
(D) Immunoblot of the eluates from affinity chromatography of COS cell cytosol using GST-fusions to Arl8b as in (A). The cells were transiently expressing full-length SKIP or SKIP(1-300) with N-terminal myc tags. The lower panel is a Coomassie-stained gel of the total material on the glutathione-Sepharose beads to indicate the levels of GST fusion proteins.
(E) Confocal micrographs of COS cells expressing the indicated versions of SKIP and stained for the myc epitope. Scale bars 10 μm.
(F) Confocal micrographs of the periphery of a COS cell expressing the full-length SKIP and costained for the lysosomal marker CD63.
(G) Confocal micrographs of COS cells cotransfected with plasmids expressing Arl8b-GFP and the indicated versions of SKIP. For SKIP(1-300) a region is enlarged in the inset panel to show the colocalization and the tubes that are particularly prominent with this truncation, although were also sometimes observed with the full-length protein. Scale bars 10 μm. See also Figure S1.