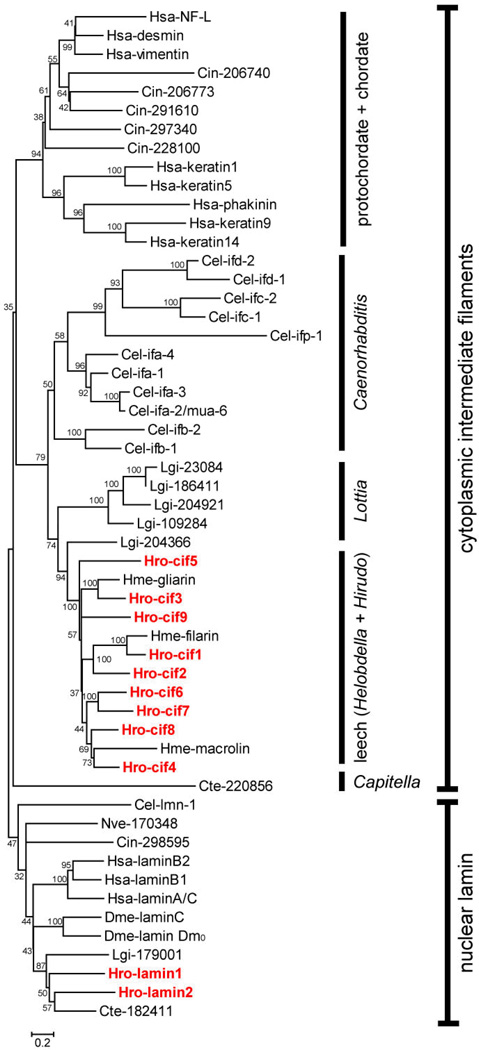
Fig. 8.
A consensus neighbor-joining tree (1000X bootstraps) of IF proteins from selected metazoan species. Helobdella IF proteins are marked in red. JGI protein ID numbers of Helobdella robusta IF proteins are given in Table 1. Proteins deduced from IF gene models in the genomes of Capitella telata (Cte), Lottia gigantea (Lgi), Ciona intestinalis (Cin), and Nematostella vectensis (Nve) are denoted with the three-letter species codes followed by their JGI protein ID numbers. Sequence and associated information of these gene models is retrievable from JGI genome portal (http://genome.jgi-psf.org/). IF proteins from Homo sapiens (Hsa), Drosophila melanogaster (Dme), Caenorhabditis elegans (Cel), and Hirudo medicinalis (Hme) are denoted with the three letter-species codes followed by specific gene names. Accession numbers: Hsa-laminA/C (NP_733821.1); Hsa-laminB1 (NP_005564.1); Hsa-laminB2 (NP_116126.2); Hsa-keratin1 (NP_006112.3); Hsa-keratin5 (NP_000415.2); Hsa-keratin 9 (NP_000217.2); Hsa-keratin 14 (NP_000517.2); Hsa-vimentin (NP_003371.2); Hsa-desmin (NP_001918.3); Hsa-NF-L (NP_006149.2); Hsa-phakinin (NP_003562.1); Dme-lamin C (NP_523742.2); Dme-lamin Dm0 (NP_476616.1); Cel-lmn-1 (NP_492371.1); Cel-ifa-1 (NP_741902.1); Cel-ifa-2/mua-6 (NP_510648.1); Cel-ifa-3 (NP_510649.3); Cel-ifa-4 (NP_508836.3); Cel-ifb-1 (NP_495136.1); Cel-ifb-2 (NP_495133.1); Cel-ifc-1 (NP_503783.1); Cel-ifc2 (NP_741705.1); Cel-ifd-1 (NP_001024830.1); Cel-ifd-2 (NP_508160.1); Cel-ifp-1 (NP_509628.1); Hme-filarin (AAD29246.1); Hme-gliarin (AAD29248.1); Hme-macrolin (AAD29247.1).