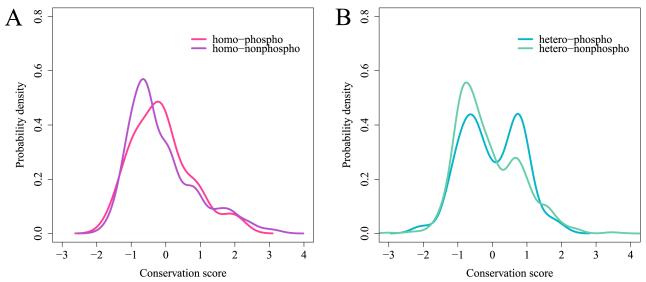
Figure 3.
Probability density function of the conservation score calculated for phosphorylation sites on binding interfaces. Zero conservation score corresponds to the same amount of evolutionary conservation as the mean conservation of the protein family. A: homooligomers, conservation of phosphorylation sites (N=275 phosphosites on interfaces) is shown in red and for non-phosphorylation sites (N=2773) in purple. B: heterooligomers, conservation of phosphorylation sites (N=521) is shown in blue and for non-phosphorylation sites (N=5559) in green. The conservation distribution for phosphosites is significantly shifted toward positive values compared to conservation of interfacial non-phosphosites for all complexes and for heterooligomers in particular (p-value = 0.00001 for all and p-value = 0.015 for heterooligomers). The distributions are smoothed by the Gaussian kernel density estimation. See also Figure S3.