Figure 2. The ESA interacts with the deacetylase core.
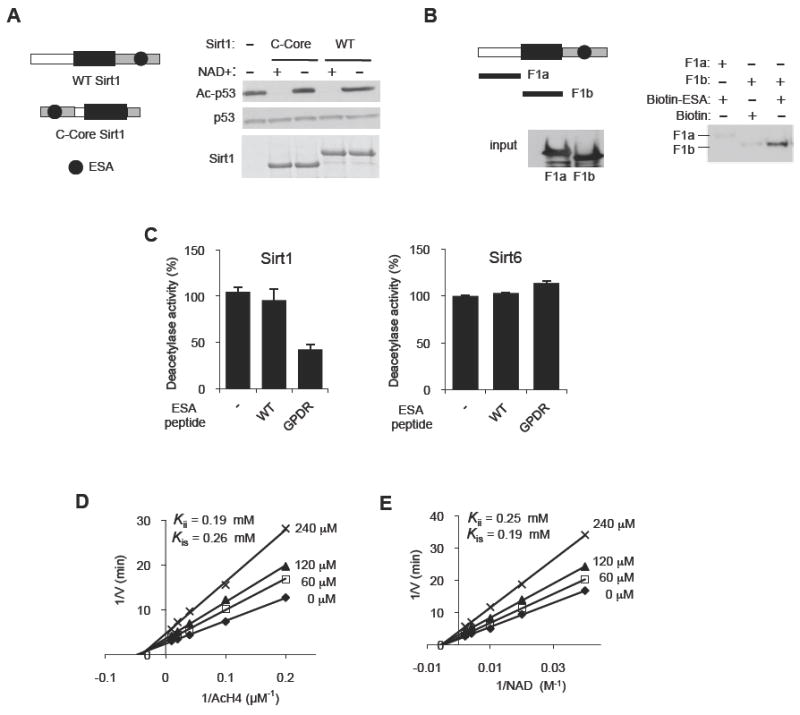
(A) The position of the C-terminal domain relative to the deacetylase core is not critical for Sirt1 deacetylase activity. Deacetylation reactions were performed with WT Sirt1 or Sirt1 in which the C-terminal domain (gray, a.a. 611-737) was placed on the N-terminal side of the deacetylase core (C-Core, a.a. 184-510). See Figure S2A-D for the disordered nature of the C-terminal domain of Sirt1.
(B) The ESA binds to the Sirtuin domain. Pull-down assays were performed after incubating biotin-tagged ESA peptide immobilized on streptavidin beads with either F1a (a.a. 1-236) or F1b (a.a. 236-490) fragments. Streptavidin-immobilized biotin without the peptide is used as negative control. F1a or F1b bound to the ESA peptide is shown above, and the input levels of F1a and F1b are shown below.
(C) The GPDR mutant ESA peptide can inhibit the deacetylase activity of Sirt1 (left panel), but not of Sirt6 (right panel), in trans. The deacetylase activity of recombinant Sirt1 and Sirt6 was measured by a fluorometric assay in the presence of 150 μM of either the WT ESA peptide, or the GPDR-mutant ESA peptide (GPDR) or no peptide (-) (N=4). Results are expressed as the mean ± s.e.m. See also Figure S2E-G.
(D, E) The GPDR-mutant ESA peptide inhibits Sirt1 in a noncompetitive manner. A Lineweaver-Burk plot derived from a fluorometric measurement of WT Sirt1 activity in the absence or presence of 0 (◆), 60 (□), 120 (▲) or 240 (×) μM of the GPDR-mutant ESA peptide for the indicated concentration range of Ac-H4 (D) and NAD+ (E). Inhibition constants (Kii or Kis) for the GPDR peptide are shown for both Ac-H4 and for NAD+ substrates. For each inhibition analysis, the concentrations of the indicated substrate and the inhibitors were varied while the concentration of the other substrate was kept constant (500 μM NAD+ for (D) and 100 μM Ac H4 for (E)). Each experiment was performed in triplicates. To calculate the inhibition constants, nonlinear mixed-effects model fitting function in the programming language R, nlme, was used for data fitting against double-reciprocal form of the noncompetitive model equation as described in the Experimental Procedures section.
