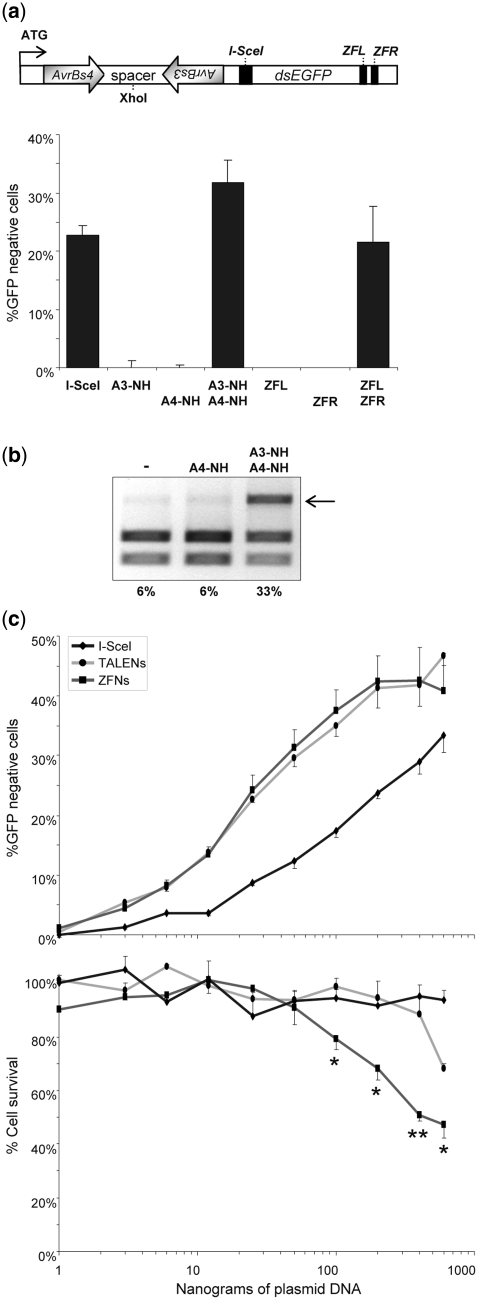
Figure 3.
Comparison of activity versus toxicity profiles of TALEN and ZFN. (a) Designer nuclease-mediated gene disruption. The HEK293-based reporter cells harbor an integrated dsEGFP gene that contains an inverted heterodimeric AvrBs4/AvrBs3 target sequence separated by 13-bp spacer in the 5′-end of the open reading frame. The position of the diagnostic XhoI site is indicated. For internal reference, a binding site for I-SceI was placed downstream of the TALEN target site. The graph shows the percentage of EGFP-negative cells 5 days after transfection with the nuclease expression vectors (TALEN, ZFN or I-SceI). (b) Molecular characterization. Genomic DNA was extracted 5 days after transfection and PCR amplicons encompassing the target sites were used as templates for digestion with XhoI. An arrow indicates the position of the XhoI-resistant DNA fragment. The numbers below designate the percentages of XhoI-resistant PCR fragments (note background level of ~6%). (c) Activity versus toxicity. The EGFP reporter cells were transfected with increasing amounts (1–600 ng) of nuclease expression vectors (TALEN, ZFN or I-SceI) and a mCherry-encoding plasmid. The percentage of EGFP and mCherry-positive cells was determined by flow cytometry after 2 and 5 days. The graphs display gene disruptions activities (EGFP-negative cells at Day 2; top) and nuclease-associated cytotoxicities (fraction of mCherry-positive cells at Day 5 as compared to Day 2 after transfection; bottom), relative to cells transfected with a mock plasmid. Statistically significant differences in toxicities between TALEN and ZFN are indicated by *P < 0.05 or **P < 0.01, respectively.