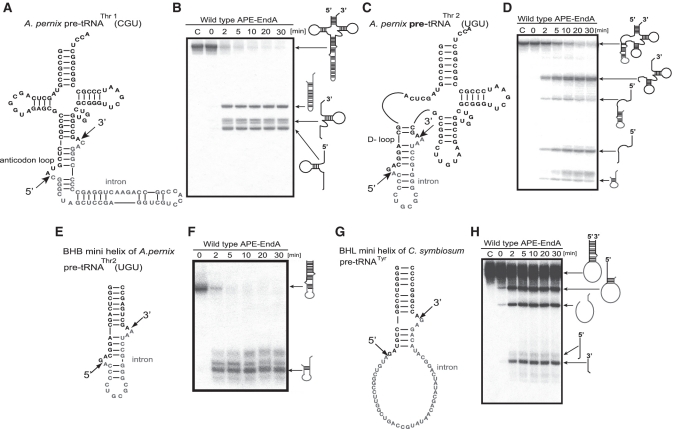
Figure 3.
Splicing activity and specificity of APE-EndA. (A) Predicted secondary structure of A. pernix pre-tRNAThr1(CGU). (B) Time-dependent cleavage of A. pernix pre-tRNAThr1(CGU). (C) Predicted secondary structure of A. pernix pre-tRNAThr2 (UGU). (D) Time-dependent cleavage of A. pernix pre-tRNAThr2(UGU). (E) Predicted secondary structure of the BHB mini helix of A. pernix pre-tRNA Thr2(UGU). (F) Time-dependent cleavage of BHB mini helix. (G) Predicted secondary structure of the BHL mini helix of C. symbiosum pre-tRNATyr. (H) Time-dependent cleavage of the BHL mini helix of C. symbiosum pre-tRNATyr. Short arrows in (A), (C), (E) and (G) indicate the splicing sites for each pre-tRNA. Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. In each gel, ‘C’ (first lane) indicates control (no enzyme). The cleavage products are shown using schematic models at the right hand side of the gel.