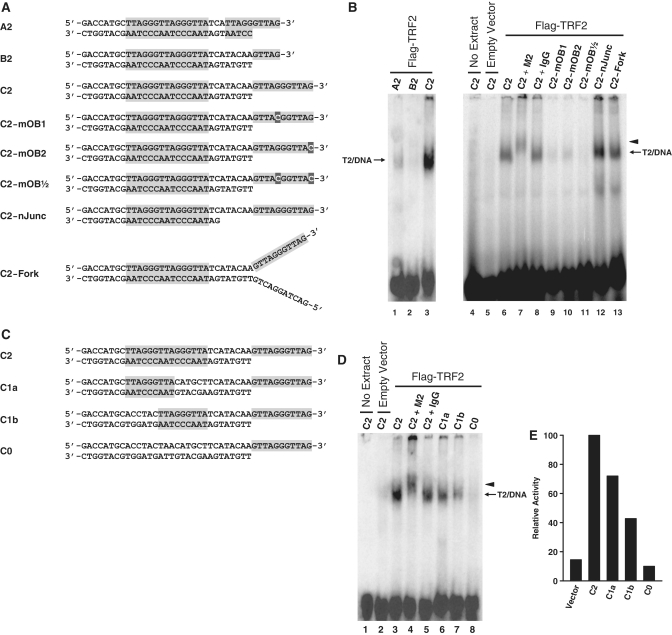
Figure 3.
Characterization of the DNA binding specificity of complex T2. (A and C) Graphical representations of the structures of all probes tested. Probes were designed to harbor different end structures and to contain different number of ds-TTAGGGTTA motifs. Shaded areas represent telomeric DNA. (B) DNA binding by complex T2 requires a functional POT1 binding site. Probes described in A were incubated with no extracts (lane 4) or with nuclear extracts of HT1080-Vector (lane 5) or HT1080-TRF2 cells (lanes 1–3 and 6–13), after which point the protein/DNA complexes were resolved by native electrophoresis in composite gels containing TAM buffer. Five minutes prior to loading, antibodies were added to samples in lanes 7 (anti-Flag M2 antibody) and 8 (Normal mouse IgG). Lanes 1–3 were overexposed to allow detection of the signal of probe A2. Short arrow indicates positions of the supershifted complex T2. (D) DNA binding by complex T2 requires at least one Myb binding site. Probes described in C were incubated with no extracts (lane 1) or with nuclear extracts of HT1080-Vector (lane 2) or HT1080-TRF2 cells (lanes 3–8), after which point the protein/DNA complexes were resolved by native electrophoresis in composite gels containing TAM buffer. Five min prior to loading, antibodies were added to samples in lanes 4 (anti-Flag M2 antibody) and 5 (Normal mouse IgG). Short arrow indicates positions of the supershifted complex T2. (E) Densitometric quantification of the T2/DNA complexes detected in (D).