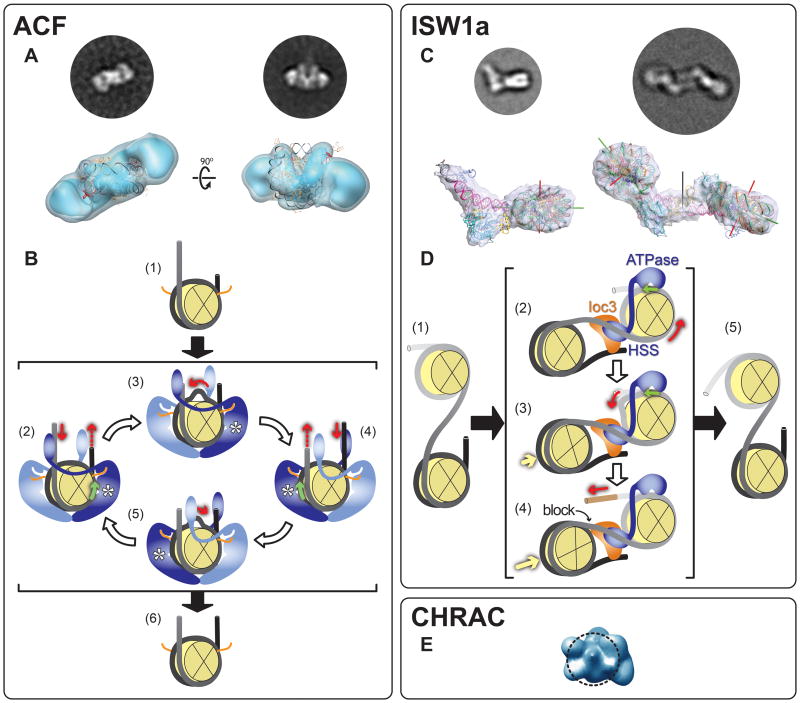
Figure 2. Electron microscopy reconstructions of ISWI family chromatin remodelers.
This figure shows reconstructions for three remodelers in the ISWI family: (A) SNF2h, the ATPase subunit of ACF (human) [17**]; (C) a version of ISW1a (S.cerevisiae) lacking the ATPase domain of Isw1, its calytic subunit [18**] and (E) CHRAC (human) [20]. The figure also illustrates mechanisms proposed for SNF2h/ACF (B) and ISW1a (D) based on structural and other biochemical data [18**]. The figure is divided into three panels, one for each type of remodeling complex. The structures of SNF2h (ACF panel) and CHRAC were obtained from negatively stained samples while those of ISW1a(ΔATPase) are from frozen-hydrated samples (Box 1). The circular images are experimental “class averages” (Box 2), corresponding to the view of the reconstruction shown below them. (A, bottom row) Two views are shown of a reconstruction consisting of two SNF2h monomers bound to a nucleosome. The atomic structure of the nucleosome (PDB 1KX5) was fitted into the density by the authors [17**]. The SNF2h monomers bind to an area surrounding the superhelical locations (SHL) −2 and +2. (B) A proposed mechanism for ACF's nucleosome centering activity (compare (1) with (6)) (adapted from [17**]). The histone octamer is depicted as a yellow sectioned disk and DNA as black/grey tubing. The orange lines represent the histone H4 N-terminal tails, which have been shown to be recognized by and modulate the activity of ISWI-type ATPases ([35-41]). SNF2h is shown in either light or dark blue, representing inactive (nucleotide-free) and active (nucleotide-bound) states, respectively. The expected location of the ATPase domain in the active monomer is marked with an asterisk. Red arrows indicate the direction of DNA motion. Green arrows mark the position, and direction, of DNA translocation within the nucleosome. (2) Two ACF monomers bind to a nucleosome; if a monomer containing ATP binds to its cognate histone H4 N-terminal tail and interacts with a longer DNA linker, its translocation activity is stimulated, leading to DNA being drawn into the nucleosome from that linker. (3) DNA diffuses through the nucleosome and emerges at the other end. (4) As the opposite linker becomes longer, the translocation activity of the second SNF2h is stimulated, resulting in translocation in the opposite direction (5). This tug-of-war between the two SNF2h monomers results in a histone octamer centered in its DNA (6). The contact between the two SNF2h monomers is hypothetical and was only included to symbolize communication between them. An alternative mechanism is discussed in [17**]. (C, bottom row) Two different structures were solved from frozen-hydrated samples containing nucleosomes and “ISW1a(ΔATPase)”, a complex consisting of the C-terminal HAND-SANT-SLIDE (HSS) portion of the ATPase Isw1 and the accessory subunit Ioc3 . In the first structure (left), the nucleosome contained DNA linkers of 45 and 29 base pairs. A density was seen bridging these two DNA extensions where ISW1a(ΔATPase) could be docked. The second structure (right) was obtained with nucleosomes containing a single 45 bp linker. In this case, a two-fold symmetric structure was observed, mediated by two copies of ISW1a(ΔATPase) that had adopted a new orientation relative to the nucleosome. The nucleosome dyad axis is shown with a green rod; its superhelical axis with a red rod and the two-fold dyad axis of the dimeric particle with a black rod. (D) The two modes of interaction between ISW1a(ΔATPase) and a nucleosome observed in the cryo-EM reconstructions as well as the interactions observed in the ISW1a(ΔATPase)-DNA co-crystal structure were used to propose a model for ISW1a's ability to take an array of randomly distributed nucleosomes (1) and evenly space them (5). The depiction of nucleosomes and the meaning of the red and green arrows are the same as in (B). Isw1 is represented in blue, with its HAND-SANT-SLIDE portion binding to linker DNA and its ATPase domain (not present in the cryo-EM reconstructions) modeled as bound at superhelical location (SHL) −2/+2, as previously mapped ([19,42,43]). The accessory Ioc3 subunit is shown in orange. The yellow arrows indicate nucleosome movement. (2) ISW1a engages its substrate with a number of interactions: the ATPase domain binds to the nucleosome where translocation will occur (the “mobile” nucleosome); the HSS portion of Isw1 binds to the DNA linking this nucleosome to the neighboring one (the “static” nucleosome, where no translocation occurs) and the Ioc3 subunit bridges this same DNA to the distal linker in the static nucleosome while at the same time interacting with HSS. (2 and 3) The Isw1 ATPase translocates DNA from the linker joining the mobile and static nucleosomes and this DNA emerges at the distal linker in the mobile nucleosome [brown segment in (4)]. Once the static nucleosome reaches the Ioc3 subunit (4) further translocation is blocked, resulting in a fixed DNA linker length between the two nucleosomes (5). (E) A view of the reconstruction of CHRAC. The dashed circle represents the contour of a nucleosome and indicates the proposed interaction between CHRAC and one of the disk-like histone faces of the nucleosome. Images adapted with permission from Macmillan Publishers Ltd: Nature (2009) [17**] and (2011) [18**].