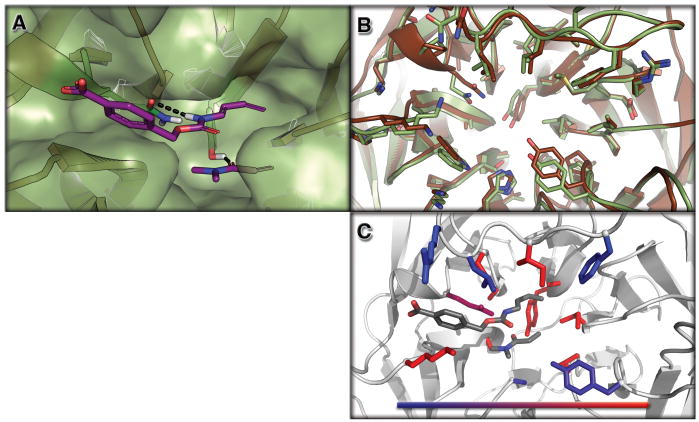
Figure 2. Structure of a Designed Diels-Alderase.
Top panel (2A): surface view of the design model (DA_20_00, green) bound to the substrates (diene and dienophile, purple). The catalytic residues making the designed hydrogen bonds are depicted as sticks. Middle panel (2A): overlay of the design model (DA_20_00, green) and the apo enzyme crystal structure of DA_20_00_A74I (brown). Bottom panel (2C): Contribution of designed residues to catalysis assessed through reversions back to the native amino acid at each position individually. Colors indicate the reduction in activity upon reversion to the native amino acid; 2 fold (blue) to > 10 fold (red). The figure was generated with PyMol (15).