Figure 1. RNA virus jelly rolls.
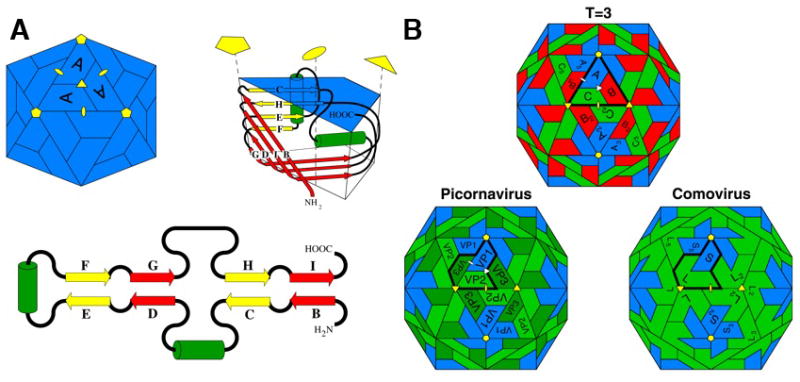
(A) A diagram of the viral jelly roll tertiary structure observed in many ssRNA icosahedral virus capsids (top right), its topology (lower) and its disposition in a T=1 icosahedral particle (top left). β-strands (represented by yellow or red arrows that correspond to their sheets in the folded structure) and positions of insertions between strands are shown in the topological representation. The green cylinders represent helices that are usually conserved. The C–D, E–F, and G–H loops often contain significant insertions. The fold is named after the procedure a baker would use to prepare a jelly roll, i.e. making an extended piece of dough, folding it back on itself, as in the topological figure, and then rolling it up as in the tertiary structure figure.
(B) The relationship between capsids formed by 180 jelly rolls arranged with icosahedral symmetry. Top: a T=3 capsid (typical of ssRNA plant viruses) formed by one type of gene product. Positions A, B, and C are occupied by subunits with identical amino acid sequences. Lower left: The picornavirus capsid has 3 different gene products, all with jelly roll folds, but without a detectable sequence similarity, occupying the positions Vp1, Vp2 and Vp3. These are equivalent, in terms of the quaternary structure, to A, C and B in the T=3 virus. Lower right: The comovirus capsid in which VP2 and VP3 are fused by a linker to form a single polypeptide chain made of two jelly rolls. Picorna virus capsids probably evolved from a T=3 capsid in which the subunit gene triplicated to form three jelly rolls that evolved independently. Eventually they were separated by post-translational proteolysis. The comovirus capsid appears to be an intermediate in this process.
