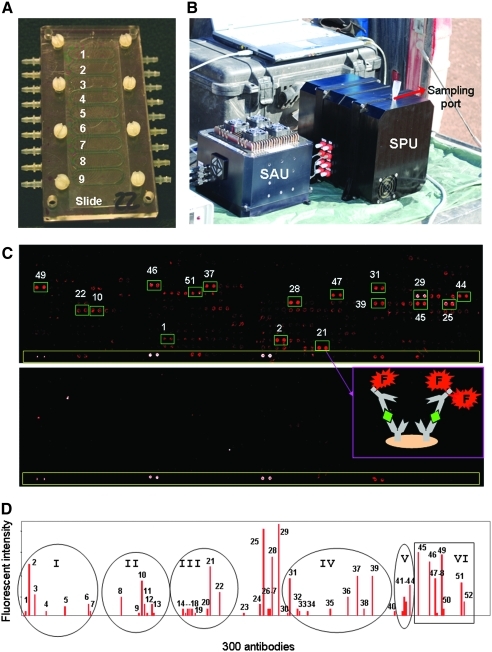
FIG. 3.
LDChip300 detected microbes and polymeric biological remains at 2 m deep under the Atacama surface. (A) A picture showing the nine flow cells for nine simultaneous LDChip300 assays in the MAAM device (see Section 2.6.1). (B) A picture of the SOLID3 instrument in the field, showing its two functional units: the Sample Preparation Unit (SPU) and the Sample Analysis Unit (SAU) (See Section 2.6.2). (C) LDChip300 image obtained with sample 200 (top) indicating some of the spots with the highest fluorescent signals. The cartoon shows how capturing antibodies (Y forms) bind biological polymers (rhombi) and how they are sandwiched by fluorescent (F) tracer antibodies. The image obtained with the negative control (bottom) was used as a baseline (blank) to calculate spot intensities in the test samples. Long yellow rectangles encompass a fluorescent spot gradient. (D) Immunogram showing the fluorescence intensity of the positive spots. The peak numbers, antibody name, and immunogen can be seen in Table 3 (see also Table 2). Circles and squares indicate the main antibody clusters: (I) antibodies against whole extracts from sediments, biofilms, and water from Río Tinto rich in Gammaproteobacteria; (II) Gammaproteobacteria (Halothiobacillus spp., Acidithiobacillus spp.); (III) Desulfosporosinus, Pseudomonas, Salinibacter; (IV) different peptides and proteins; (V) cell wall polymeric components like lipoteichoic acids, lipidA, or peptidoglycan; (VI) nitroaromatic compounds and nucleotide derivatives. Color images available online at www.liebertonline.com/ast