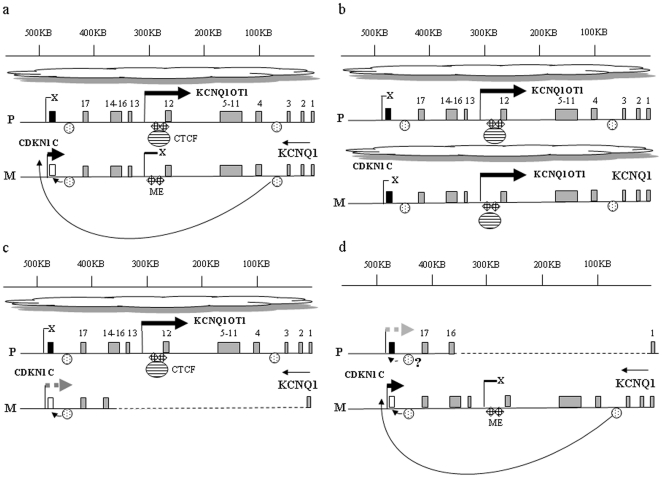
Figure 4. Model of normal and disrupted imprinting regulation at the CDKN1C locus.
Genomic distances were calculated from NCBI reference sequence NG_008935.1. (a) CDKN1C imprinting is maintained on the paternal chromosome by KCNQ1OT1 transcription leading to bidirectional gene silencing (indicated by spreading cloud) and formation of a chromatin insulator within the KVDMR (CTCF binding) that prevents a dominant distant CDKN1C enhancer from activating paternal CDKN1C expression. On the maternal chromosome, methylation within the KVDMR prevents chromatin insulator formation, KCNQ1OT1 is silent, and the distance enhancer can activate maternal CDKN1C expression. (b). In BWS cases with LOM within KvDMR, bidirectional gene silencing on both paternal and maternal chromosomes occurs, chromatin insulators exist on the maternal and paternal chromosomes and distant CDKN1C enhancers can no longer activate maternal CDKN1C expression. Mosaicism for the LOM epimutation allows some CDKN1C expression to occur. (c). Maternal deletion of 330 kb within the KCNQ1 locus removes the KvDMR, and the distant enhancer element, thereby reducing the maternal CDKN1C expression that is normally activated by distant enhancers. Enhancers located close to CDKN1C, not affected by deletion, may still be active and allow CDKN1C expression in some tissues. (d). Paternal deletion of 330 kb has a minimal effect on CDKN1C expression because even though the KvDMR is deleted on the paternal chromosome, the distant enhancer for CDKN1C is also deleted, neutralizing any potential for activation of paternal CDKN1C. Proximal CDKN1C enhancers unaffected by deletion may be active in some tissues. Maternal CDKN1C expression is unaffected.