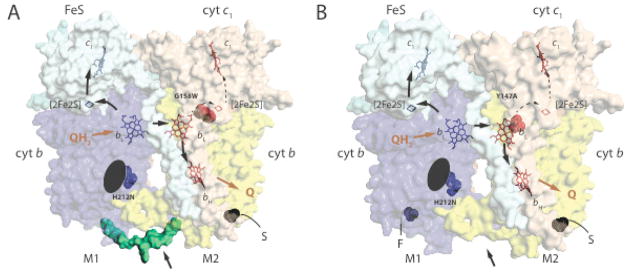
Fig. 1. Structure and function of heterodimeric cytochromes bc1.
A. A hypothetical structural model for a non-native heterodimeric cytochrome b-bc1. The 3D structure of cytochrome bc1 (PDB entry 1ZRT) was modified to include the Qi site H212N mutation (blue) on one monomer (M1) and the Qo site G158W mutation (red) on the other (M2) to depict a heterodimeric enzyme produced by the one-plasmid system [29]. The carboxyl end of one cytochrome b (blue) is linked to the amino end of the other (yellow) with a peptide (green) indicated by an arrow to create cytochrome b-b that is not naturally present in cytochrome bc1. S refers to the Strep–tag on the carboxyl terminal end of cytochrome b-b. The Fe/S and cytochrome c1 subunits are shown in light blue and light brown, with the cofactors ([2Fe2S] and hemes bH, bL and c1) in monomer M1 in blue and those in M2 in red. The black arrows indicate the electron transfer pathways across the monomers, Q and QH2 correspond to quinone and hydroquinone, respectively. The black ellipsoid indicates the absence of heme bH on monomer M1. B. A hypothetical structural model for a native-like heterodimeric cytochrome bc1. The 3D structure of cytochrome bc1 (PDB entry 1ZRT) was modified to include the Qi site H212N mutation (blue) on one monomer (M1) and the Qo site Y147A mutation (red) on the other (M2) to depict a heterodimeric enzyme produced by the two-plasmids system [32]. The labels are as in A, and note the absence (indicated by an arrow) of the peptide linking together the cytochromes b, and the presence of Flag- (F) and Strep (S)-tags on the carboxyl terminals of two cytochrome b on monomers M1 and M2, respectively.