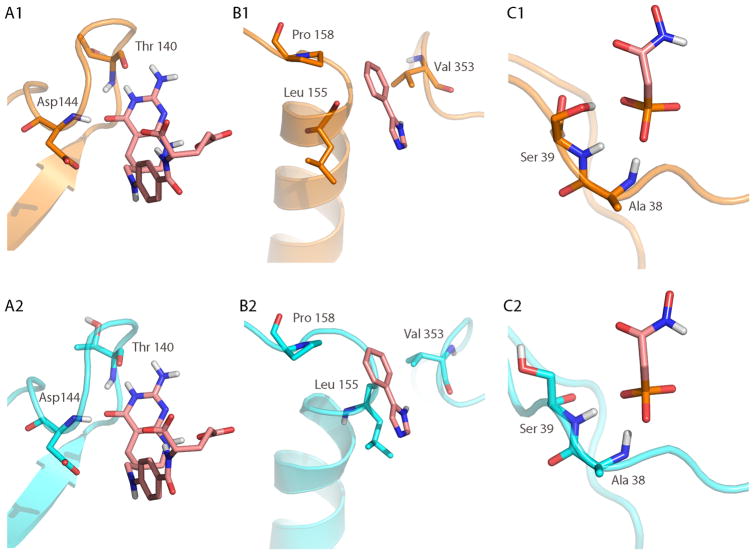
Figure 6.
Comparison of loop-ligand interactions between the X-ray structure (top row, shown in orange) and the lowest RMSD predicted structure (bottom row, shown in cyan). The co-crystallized ligand is shown in pink sticks in all panels. (A2) In the predicted loop structure of GART, the ligand forms the same two hydrogen bonds (Thr 140 and Asp 144) with the backbone of the loop as seen in the X-ray structure (A2). In CYP119, hydrophobic contacts dominate the interaction with the ligand. The contacts to Val 353 and Pro 158 are well conserved between the X-ray (B1) and predicted structure (B2). The hydrophobic contact with Leu 155, although present, has sterically changed in the predicted structure compared to the X-ray structure. (C) In the enolase system, the phosphate atom of the ligand interacts with two backbone amides (Ala 38 and Ser 39), but loses a hydrogen bond contact to the side chain of Ser 39 in the predicted structure.