Figure 1.
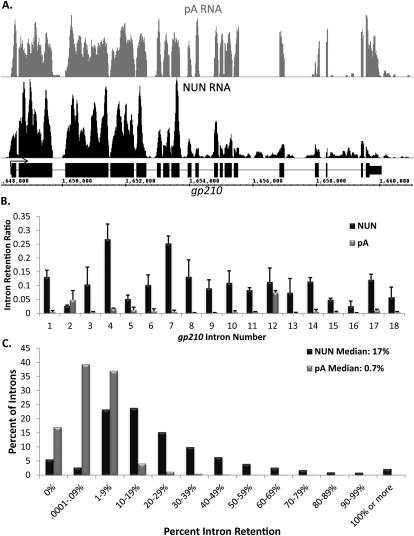
Most introns in Drosophila S2 cells are efficiently spliced cotranscriptionally. (A) An image of the sequencing reads for a typical gene, gp210, in the Affymetrix Integrated Genome Browser. pA RNA is in gray, NUN RNA is in black, and gene structure is in black. Note breaks in sequencing reads coinciding with intron position. (B) Quantitation of intron retention for the introns of gp210. Intron retention = reads per base pair in introns/reads per base pair in all exons. (C) A histogram of the percent of all introns of abundantly transcribed genes, grouped by intron retention. pA RNA is in gray, and NUN RNA is in black.