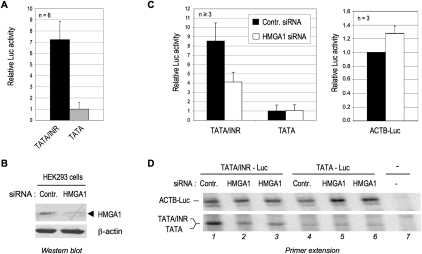
Figure 4.
HMGA1 is required selectively for TATA/INR but not TATA-mediated transcription in vivo. (A) TATA/INR-Luc and TATA-Luc were transfected in HEK293 cells, and the relative luciferase activities (mean ± SD) from six independent experiments (each in duplicate) are shown. The luciferase activity of TATA-Luc was arbitrarily set to 1. (B) HEK293 cells were transfected with either control (Contr.) or specific siRNA against HMGA1. Endogenous HMGA1 in whole-cell extracts was analyzed by Western blot. (C) TATA/INR-Luc, TATA-Luc, or ACTB-Luc reporters were transfected in HEK293 cells with a control siRNA (black bars) or the HMGA1-specific siRNA (open bars). The relative luciferase activities are shown (as in A). TATA-Luc activity (left panel) and ACTB-Luc activity (right panel) in cells transfected with the control siRNA were set arbitrarily to 1. (D) Total mRNA from HEK293 cells transfected with ACTB-Luc (lanes 1–6) and either TATA/INR-Luc (lanes 1–3) or TATA-Luc (lanes 4–6) and the indicated siRNAs was analyzed by primer extension with the Luc 24mer primer. Lane 7 is a control reaction with mRNA from mock-transfected cells. The position of correctly initiated transcripts is indicated for each promoter construct (length of 172 nucleotides [nt] for ACTB-Luc and 128 nt for TATA and TATA/INR-Luc). The top and bottom autoradiograms are from the top and bottom parts of the same gel; the bottom autoradiogram is from a longer X-ray film exposure time. See also Supplemental Figure S4 for an analysis of a different TATA/INR core promoter.