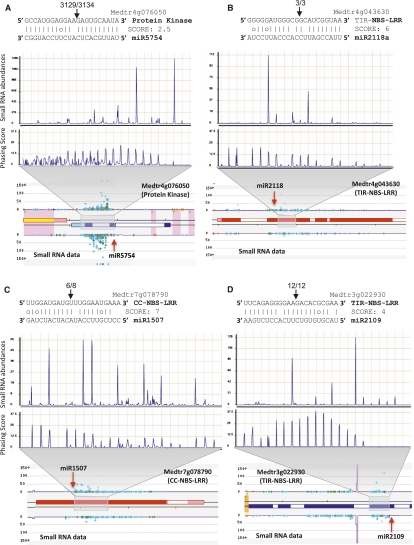
Figure 1.
Twenty-two-nucleotide miRNAs trigger phased siRNA production in M. truncatula. Above are alignments of well-conserved miRNAs and their targets; in the alignment, vertical lines indicate matches, missing lines indicate mismatches, and G:U wobble pairs are indicated with a circle. Black arrowheads above the target sequence indicate the cleavage site in the target, and the numbers above separated by the backslash indicate the number of PARE reads in the small window (WS) (first number) and the number of reads in the large window (WL) (second number), as described in the Supplemental Material. Below the alignments are small RNA abundances and phasing score distributions for the regions indicated by the gray trapezoids; abundances are normalized in TPM (transcripts per million). Below this are images from our Web site showing the M. truncatula flower small RNA data at each PHAS locus, with the red arrowhead pointing to the miRNA cleavage site. Colored spots are small RNAs with abundances indicated on the Y-axis; light-blue spots indicate 21-nt sRNAs, green spots are 22-nt sRNAs, orange spots are 24-nt sRNAs, and other colors are other sizes. Blue boxes (on the bottom strand) or red boxes (on the top strand) are annotated exons. Purple lines indicate a k-mer frequency for repeats; yellow, pink, or orange shading indicates DNA transposons, retrotransposons, or inverted repeats. (A) The new miRNA mtr-miR5754 targets a gene encoding a protein kinase; both the miRNA trigger and the phasiRNAs are specific to the flower library. (B) miR2118 targets a gene encoding a TNL. (C) miR1507 targets a gene encoding a CNL. (D) miR2109 targets a gene encoding a TNL.