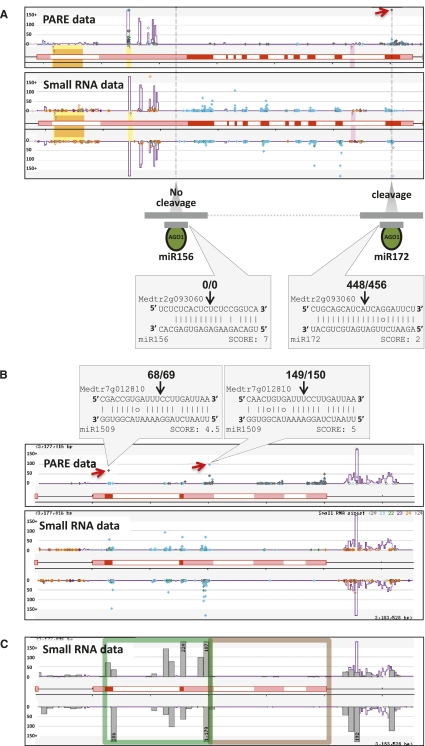
Figure 2.
Novel classes of tasiRNAs identified in the M. truncatula genome. (A) Example of a 221 TAS locus that encodes an AP2 homolog (Medtr2g093060). The top panel shows the PARE data with a high-abundance tag from the cleaved site (red arrowhead); for space reasons, only the coding strand data are shown for the PARE tags. The image is interpreted as described in Figure 1. The small RNA data are below; colored dots indicate small RNA sizes, with light blue indicating 21-mers. Other features are as described for the PARE images. The bottom section illustrates the predicted noncleaving miR156 site and the cleaved miR172 site, along with alignments of those miRNAs with the AP2 transcript and the PARE tag abundances. (B) An example of 222 TAS locus (Medtr7g012810); double cleavage by the 22-nt miRNA miR1509 triggers phasiRNAs. The first cleavage site occurs on Chr. 7 at nucleotide position 3,178,284; the second is at position 3,180,023. Both cleavage sites and alignments are indicated, as in A. (C) The small RNA data from the bottom panel of B as a histogram of summed sRNA abundances to emphasize the increased abundance between the two cleavage sites (red arrows in the top panel of B). The green box outlines the region between the two cleavage sites; the brown box indicates the region 3′ of the 3′ 22-nt miRNA cleavage site to the last gene-associated small RNA.