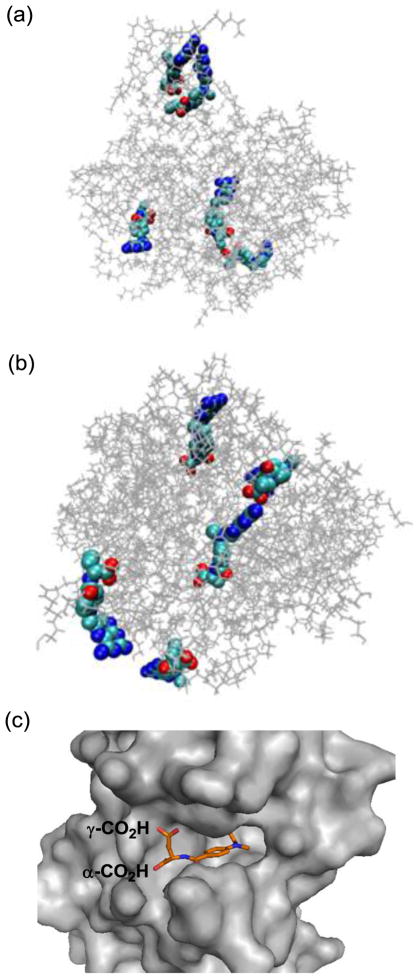
Figure 5.
(a, b) Molecular models generated by Molecular Dynamics simulation for the representative members of generation 5 PAMAM dendrimer conjugated with methotrexate, either directly via an amide bond (2c, Scheme 1) or through a medium length of spacer (7a, Scheme 2). Distribution of MTX molecules on each of the dendrimers was selected arbitrarily for this simulation purpose, and thus should be considered as one of the potential distributions. Final configurations for 2c (a) and 7a (b) were acquired after 1-ns MD simulations. The figures of the structures were generated with the software VMD (visual molecular dynamics) where the PAMAM dendrimer and MTX were shown as lines and van der Waals surface, respectively (Colors: grey: dendrimer, light blue: carbon, blue: nitrogen, red: oxygen); (c) A crystal structure of human dihydrofolate reductase (hDHFR) in complex with a methotrexate molecule at its active site (PDB code 1u72), where the L-glutamate carboxylic acids from the drug molecule are anchored near the entrance to the enzyme catalytic pocket, while a pteridine head group (hidden) is bound deep into the pocket.