Abstract
In this paper, we introduce a novel knowledge-based word sense disambiguation method that determines the sense of an ambiguous word in biomedical text using semantic similarity or relatedness measures. These measures quantify the degree of similarity between concepts in the Unified Medical Language System (UMLS). The objective of this work was to develop a method that can disambiguate terms in biomedical text by exploiting similarity information extracted from the UMLS and to evaluate the efficacy of information content-based semantic similarity measures, which augment path-based information with probabilities derived from biomedical corpora. We show that information content-based measures obtain a higher disambiguation accuracy than path-based measures because they weight the path based on where it exists in the taxonomy coupled with the probability of the concepts occurring in a corpus of text.
Introduction
Word Sense Disambiguation (WSD) is the task of automatically identifying the appropriate sense (or concept) of an ambiguous word based on the context in which the word is used. In our work, the set of possible meanings for a word are the Concept Unique Identifiers (CUIs) associated with a particular term in the Unified Medical Language System (UMLS). Thus when performing WSD of biomedical terms, our more specific goal is to assign a term one of its possible CUIs based on its surrounding context. For example, the term cold could refer to the temperature (C0009264) or the common cold (C0009443), depending on the context in which it occurs.
Automatically identifying the intended sense of ambiguous words improves the performance of clinical and biomedical applications such as medical coding and indexing for quality assessment, cohort discovery and other secondary uses of data. These capabilities are becoming essential tasks due to the growing amount of information available to researchers, the transition of US health care documentation towards electronic health records, and the push for quality and efficiency in healthcare.
In this paper, we introduce UMLS::SenseRelate, a novel knowledge-based WSD method that disambiguates terms in biomedical text. This method determines the most context-appropriate sense of an ambiguous word using the degree of semantic similarity between the possible senses and the terms surrounding the ambiguous word. The underlying assumption of the algorithm is that the ambiguous word will be used in the sense that is most similar to the sense of the terms that surround it. We evaluate our method on path-based and information-content (IC) based similarity measures. Path-based measures rely on the hierarchical relations between the terms in a taxonomy. IC-based measures augment this information with probabilities derived from a corpus of text. IC quantifies the specificity of a concept in a hierarchy; a concept with a high IC value is more specific to a topic than one with a low IC value.
The objective of this work is two-fold. Our first objective is to develop and evaluate a method that can disambiguate terms in biomedical text by exploiting similarity information extrapolated from the UMLS. Our second objective is to evaluate the efficacy of IC-based semantic similarity measures over path-based measures.
Background
Unified Medical Language System:
The UMLS is a data warehouse containing three knowledge sources: the Metathesaurus, the Semantic Network and the SPECIALIST Lexicon. The Metathesaurus contains approximately 1.7 million biomedical and clinical concepts from over 100 different terminologies that have been semi-automatically integrated into a single source. The terminologies in the Metathesaurus can be treated independently or in combination. The Metathesaurus contains two main types of hierarchical relations between the concepts: parent/child (PAR/CHD), which are hierarchical relations between concepts that have been explicitly defined by the terminology, and broader/narrower (RB/RN), which are created by the UMLS editors during the integration process. In our experiments, we use the Medical Subject Heading (MSH) Thesaurus which is the National Library of Medicine’s (NLM) controlled vocabulary thesaurus consisting of biomedical concepts created for the purposes of indexing. The MSH terms are organized in a hierarchical structure in order to permit searching at various levels of specificity.
The Semantic Network consists of a set of broad subject categories called semantic types in which each concept in the Metathesaurus is assigned one or more semantic type. For example, the semantic type of C0206250 [Autonomic nerve] is Body Part, Organ, or Organ Component. The SPECIALIST Lexicon contains terms that are used in the biomedical and health-related domain along with linguistic information such as spelling variants. In this work, we use the SPECIALIST Lexicon to identify terms surrounding the ambiguous word in our dataset.
Medline:
Medline a is a bibliographic database containing over 18.5 million citations to journal articles in the biomedical domain and is maintained by NLM. The 2009 Medline Baseline encompasses approximately 5,200 journals starting from 1948 and contains 17,764,826 citations; consisting of 2,490,567 unique unigrams (single words) and 39,225,736 unique bigrams (two-word sequences). The majority of the publications are scholarly journals but a small number of newspapers and magazines are included.
Related Work in Biomedical WSD
Existing methods that have been proposed to automatically disambiguate words in biomedical text can be classified into four groups: supervised,1 2 3 4 5 semi-supervised,6 unsupervised,7 and knowledge-based methods.8 9 Supervised and semi-supervised methods use machine learning algorithms to assign senses to instances containing the ambiguous word. These algorithms learn from annotated training data which consists of a sufficient number of instances for each sense of an ambiguous word. Supervised methods use manually annotated training data containing instances of a the ambiguous word (referred to as the target word) to learn the context in which target words are used where semi-supervised methods automatically create these data. The sense inventory used in these methods are embedded in the training data. The disadvantage of these types of methods is that training data needs to be created for each target word to be disambiguated. Whether this is done manually or automatically, it is infeasible to create such data on a large scale.
Knowledge-based methods do not use any manually or automatically generated training data, but use information from an external knowledge source and possibly a corpus of text. The sense inventory for these methods comes from the knowledge source being used. Unsupervised methods rely solely use distributional characteristics of an outside corpus and do not rely on sense information or a knowledge source. In this work, we focus on knowledge-based methods.
Humphrey et al.8 introduce a knowledge-based method that assigns a sense to a target word by first identifying its semantic type with the assumption that each possible sense has a distinct semantic type. A semantic type (st-) vector is created for the semantic type of each possible sense using one word terms in the UMLS that have been assigned that semantic type. A target word (tw-) vector is created using the words surrounding the target word. The cosine of the angle between the tw-vector and each of the st-vectors is calculated and the sense whose st-vector is closest to the tw-vector is assigned to the target word. In contrast, Alexopoulou et al.9 introduce their “Closest Sense” method which calculates the average shortest distance between the semantic type of a possible sense and the semantic types each of the words surrounding the target word. This is done for each possible sense, and the sense with the shortest distance is assigned to the target word. The limitation to each of these methods is that they rely on the semantic types of the possible senses to be distinct. Therefore, if two possible senses have the same semantic type neither of these methods is able to distinguish between them. For example, the term cortices can refer to either the cerebral cortex (C0007776) or the kidney cortex (C0022655); each with the semantic type “Body Part, Organ, or Organ Component”. Analysis of the 2009 Medline data b shows that there are 1,072,902 terms in Medline that exist in the UMLS of which 35,013 are ambiguous and 2,979 have two or more senses with the same semantic type. This indicates that approximately 12% of the ambiguous words cannot be disambiguated using the knowledge-based methods discussed above and another method is required. Our method does not have this limitation.
Similarity Measures
Existing semantic similarity measures can be categorized into two groups: path-based and information content (IC)-based. Path-based measures rely on the shortest path information, whereas IC-based measures incorporate the probability of the concept occurring in a corpus of text.
Path-based:
Rada et al.10 introduces the conceptual distance measure which is the length of the shortest path between two concepts (c1 and c2) in MSH using RB/RN relations. Caviedes & Cimino11 later evaluated this measure using the PAR/CHD relations. The path measure is a modification of this and is calculated as the reciprocal of the length of the shortest path.
Wu and Palmer12 extend this measure by incorporating the depth of the LCS. In this measure, the similarity is twice the depth of the two concepts LCS divided by the product of the depths of the individual concepts as defined in Equation 1.
| (1) |
Leacock and Chodorow13 extend the path measure by incorporating the depth of the taxonomy. Here, the similarity is the negative log of the shortest path between two concepts divided by twice the total depth of the taxonomy (D) as defined in Equation 2.
| (2) |
Nguyen and Al-Mubaid14 incorporate both the depth and LCS in their measure. In this measure, the similarity is the log of two plus the product of the shortest distance between the two concepts minus one and the depth of the taxonomy (D) minus the depth of the concepts LCS (d) as defined in Equation 3. Its range depends on the depth of the taxonomy.
| (3) |
IC-based:
IC is formally defined as the negative log of the probability of a concept. Resnik15 modified IC to be used as a similarity measure. He defined the similarity of two concepts to be the IC of their least common subsumer (LCS) as shown in Equation 4.
| (4) |
Jiang and Conrath16 and Lin17 extended Resnik’s IC-based measure by incorporating the IC of the individual concepts. Lin defined the similarity between two concepts by taking the quotient between twice the IC of the concepts’ LCS and the sum of the IC of the two concepts as shown in Equation 5. This is similar to the measure proposed by Wu & Palmer; differing in the use of IC rather than the depth of the concepts.
| (5) |
Jiang and Conrath defined the distance between two concepts to be the sum of the IC of the two concepts minus twice the IC of the concepts’ LCS. We modify this measure to return a similarity score by taking the reciprocal of the distance as shown in Equation 6.
| (6) |
Method
UMLS::SenseRelate c is a freely available open source Perl package developed to assign UMLS concepts to ambiguous terms in biomedical text. In this method, each possible sense of a word is assigned a score by summing the similarity between it and the terms surrounding the ambiguous word in a given window of context. The sense with the highest score is assigned to the target word. We identify the terms surrounding the target word using the SPECIALIST Lexicon. The sequence of words with the longest match to the terms that exist in the lexicon are treated as a single term. Once the terms are identified, the algorithm computes the similarity between the possible sense of the target word and each of the surrounding terms using the freely available open source Perl package UMLS::Similarity d developed to calculate the similarity or relatedness between biomedical terms.
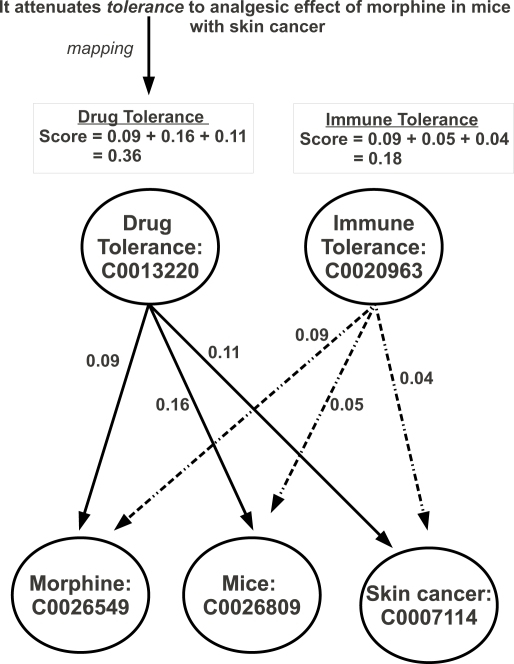
For example, consider the following sentence containing the target word tolerance which has the possible senses Drug Tolerance [C0013220] and an Immune Tolerance [C0020963]: It attenuates tolerance to analgesic effect of morphine in mice with skin cancer.
In this example, we use a window size of five which refers to five content terms to the right and the left of the target word and attempt to map them to CUIs. In this case, the content words are: attenuates, analgesic, effect, morphine, mice, skin cancer. Of these six words, only three have mappings to CUIs in MSH: morphine:C0026549, mice:C0026809, and skin cancer:C0007114. In this method, we treat skin cancer as a single term mapping to the concept C0007114 rather than individual words which would map to skin:C1123023 and cancer:C0006826.
The WSD algorithm then obtains similarity scores between each of the possible senses and the concepts of the content words in the window of context and sums the scores to obtain a total score for each possible sense as shown in Figure 1. The sense with the highest score is assigned to the target word; in this case Drug Tolerance.
Figure 1:
Example of UMLS::SenseRelate Method
As stated above, the UMLS::Similarity package is used to obtain the similarity between two biomedical terms. In previous work,18 we showed UMLS::Similarity could reliably reproduce the path-based similarity measures proposed by Leacock & Chodorow, Wu & Palmer, and Nguyen & Al-Mubaid. Subsequently, we have extended this package to include the IC-based measures proposed by Resnik, Jiang & Conrath, and Lin.
UMLS::SenseRelate is a novel extension of WordNet::SenseRelate::TargetWord developed by Patwardhan et al.19 which disambiguates words in general English text. WordNet::SenseRelate::TargetWord differs from our method in two significant aspects. The first aspect is that WordNet::SenseRelate::TargetWord is designed to disambiguate words in general English using the lexical resource WordNet which does not contain sufficient biomedical terminology20. The second aspect is UMLS::SenseRelate’s approach to addressing the identification of terms (or compound words). In the biomedical domain, many of the words surrounding the ambiguous word are predominately part of a larger term whose meaning may differ from its components. For example, Patient Controlled Analgesia can be understood by taking the union of the meanings of the three terms but the similarity between it and the word pain can not be determined by summing the similarity of its parts. In WordNet::SenseRelate::TargetWord the compounds are identified based on the lexical entries in WordNet. In our method, the terms are identified independently, in this case the SPECIALIST Lexicon, allowing for the flexibility of including outside terminology resources.
Data
Propagation Data:
The UMLSonMedline dataset created by NLM consists of concepts from the 2009AB UMLS and the number of times they occurred in a snapshot of Medline taken on 12/01/2009. The frequency counts were obtained by using the Essie Search Engine21 which queried Medline with normalized strings from the 2009AB MRCONSO table in the UMLS. The frequency of a CUI was obtained by aggregating the frequency counts of the terms associated with the CUI to provide a rough estimate of its frequency. The IC measures use this information to calculate the probability of a concept.
Evaluation Data:
We evaluate our method on NLM’s MSH-WSD dataset22. The data set contains 203 ambiguous terms and acronyms from the 2010 Medline baseline. Each instance of a term was automatically assigned a CUI from the 2009AB version of the UMLS by exploiting the fact that each instance in Medline is manually indexed with Medical Subject Headings in which each heading has an associated CUI. For each instance, containing an ambiguous word, the sense was determined by first identifying the possible CUIs of the ambiguous word in the UMLS, and second extracting the manually assigned CUIs by the indexers. If one, and only one, of the possible CUIs is in the set of manually assigned CUIs, then that CUI is assigned to the target word. Heuristic filters and manually spot checking were also conducted to ensure the dataset’s reliability. Each target word contains approximately 187 instances, has 2.08 possible senses and has a 54.5% majority sense. Out of 203 target words, 106 are terms, 88 are acronyms, and 9 have possible senses that are both acronyms and terms. For example, the target word cold has the acronym Chronic Obstructive Airway Disease as a possible sense, as well as the term Cold Temperature. The total number of instances is 37,888.
Experiment
In this paper, we evaluate each of path-based and IC-based semantic similarity measures previously discussed on the task of WSD using UMLS::SenseRelate. During this process, we also evaluate two parameters: 1) the use of terms versus single words surrounding the ambiguous word, and 2) the size of the window in which the terms and words are obtained. These experiments were conducted using the 2009AB version of the UMLS to coincide with the UMLSonMedline in which the propagation information was obtained. We use the MSH taxonomy located in the UMLS Metathesaurus because the possible senses of each of the target words in the MSH-WSD dataset were obtained from this source. Differences between the means of disambiguation accuracy produced by various approaches were tested for statistical significance using pair-wise Student’s t-test.
Results and Discussion
Table 1 shows the accuracy of UMLS::SenseRelate using the path measure (path), the path-based measures proposed by Leacock & Chodorow (lch), Wu & Palmer (wup) and Nguyen & Al-Mubaid (nam), and the IC-based measures proposed by Resnik (res), Jiang & Conrath (jcn) and Lin (lin) using various window sizes. Term refers to using the surrounding terms and Word refers to using the surrounding words.
Table 1:
Accuracy of UMLS::SenseRelate on MSH-WSD
| path | lch | wup | nam | res | jcn | lin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Window | Term | Word | Term | Word | Term | Word | Term | Word | Term | Word | Term | Word | Term | Word |
| 0 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| 1 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 |
| 2 | 0.63 | 0.59 | 0.63 | 0.59 | 0.64 | 0.59 | 0.64 | 0.59 | 0.64 | 0.60 | 0.65 | 0.61 | 0.65 | 0.61 |
| 5 | 0.66 | 0.62 | 0.66 | 0.62 | 0.67 | 0.63 | 0.67 | 0.63 | 0.68 | 0.64 | 0.68 | 0.65 | 0.69 | 0.65 |
| 10 | 0.69 | 0.64 | 0.68 | 0.64 | 0.69 | 0.65 | 0.69 | 0.65 | 0.70 | 0.66 | 0.71 | 0.67 | 0.71 | 0.67 |
| 25 | 0.71 | 0.66 | 0.69 | 0.65 | 0.70 | 0.67 | 0.71 | 0.66 | 0.73 | 0.68 | 0.73 | 0.68 | 0.74 | 0.69 |
| 50 | 0.72 | 0.66 | 0.69 | 0.65 | 0.70 | 0.66 | 0.72 | 0.67 | 0.73 | 0.69 | 0.74 | 0.69 | 0.74 | 0.70 |
| 60 | 0.72 | 0.67 | 0.69 | 0.64 | 0.70 | 0.66 | 0.72 | 0.67 | 0.73 | 0.69 | 0.74 | 0.69 | 0.74 | 0.70 |
| 70 | 0.72 | 0.67 | 0.69 | 0.64 | 0.70 | 0.65 | 0.72 | 0.67 | 0.73 | 0.69 | 0.74 | 0.69 | 0.74 | 0.70 |
The results show that IC-based measures consistently obtain a statistically significantly higher accuracy than the path-based measures (p ≤ 0.02). The lin measure obtains the highest disambiguation accuracy (p ≤ 0.01) over each of the window sizes, although the difference is not statistically significant with the measure proposed by Jiang & Conrath, it is with the measure proposed by Resnik (p ≤ 0.05).
The results also show that using the surrounding terms (Term) rather than the words (Word) obtains a statistical significantly higher disambiguation accuracy for each of the measures and window sizes (p ≤ 0.01). We believe that this is because the terms are less ambiguous than words and provide a more specific distinction. For example, when skin and cancer are individually mapped to the concepts Skin [C1123023] and Cancer [C0006826] separately, their combination does not provide the exact meaning of the concept Skin Cancer [C0007114].
We also compare the results to the majority sense baseline which is often used to evaluate supervised learning algorithms and indicates the accuracy that would be achieved by assigning the most frequent sense to every instance. The overall majority sense baseline for the MSH-WSD dataset is 0.5448. The results in Table 1 show that for each measure, the disambiguation accuracy is statistically significantly greater than the baseline (p ≤ 0.01).
The possible senses of the target words in the MSH WSD dataset can be grouped into three categories: terms (MSHWSD TERMS), acronyms (MSH-WSD ACRONYMS) and a combination (MSH-WSD TERMS/ACRONYMS). Table 2 shows the number of instances for each category, the overall accuracy for each measure when using a window size of 50 and surrounding terms (Term), and the majority sense baseline. The results show that UMLS::SenseRelate obtains a higher overall accuracy when disambiguating acronyms than terms or their combination
Table 2:
Breakdown of Results using Terms and a Window Size of 50
| Category | # instances | baseline | path | lch | wup | nam | res | jcn | lin |
|---|---|---|---|---|---|---|---|---|---|
| MSH-WSD TERMS | 88 | 0.55 | 0.64 | 0.62 | 0.64 | 0.65 | 0.66 | 0.67 | 0.67 |
| MSH-WSD ACRONYMS | 106 | 0.54 | 0.78 | 0.75 | 0.76 | 0.79 | 0.79 | 0.80 | 0.80 |
| MSH-WSD TERMS/ACRONYMS | 9 | 0.53 | 0.68 | 0.66 | 0.67 | 0.69 | 0.75 | 0.71 | 0.73 |
| MSH-WSD OVERALL | 203 | 0.54 | 0.72 | 0.69 | 0.70 | 0.72 | 0.73 | 0.74 | 0.74 |
Table 3 shows the individual results for the top five target words with the highest and lowest accuracy obtained by UMLS::SenseRelate using the lin measure, a window size of 50 and surrounding terms (Term), as well as the majority sense baseline e.
Table 3:
Top Five and Bottom Five Target Words using Lin with a Window Size of 50
| Bottom 5 | Top 5 | ||||
|---|---|---|---|---|---|
| Target Word | Accuracy | Baseline | Target Word | Accuracy | Baseline |
| hemlock | 0.40 | 0.74 | pcb | 1.00 | 0.78 |
| heregulin | 0.42 | 0.57 | hps | 0.98 | 0.56 |
| lawsonia | 0.43 | 0.86 | ccd | 0.97 | 0.70 |
| tomography | 0.48 | 0.50 | rsv | 0.96 | 0.74 |
| ca | 0.49 | 0.25 | mcc | 0.96 | 0.76 |
The results show that all of the top five target words are acronyms. We believe that this is because, in general, the contextual distinction between acronyms is more coarse grained. Although, the target word ca scored in the bottom five indicating that this is not always the case. The target word ca has four possible senses: 1) Calcium [C0006675], 2) California [C0006754], 3) Canada [C0006823] and 4) Hippocampus [C0019564]. Table 4 shows the confusion matrix for the results of ca. The number 2 in the cell California/Canada indicates that two instances in which CA referred to California were labeled as Canada by our method.
Table 4:
Error Analysis of the Target Word CA using Lin with a Window Size of 50
| Calcium C0006675 | California C0006754 | Canada C0006823 | Hippocampus C0019564 | |
|---|---|---|---|---|
| Calcium [C0006675] | 52 | 47 | ||
| California [C0006754] | 31 | 31 | 2 | 35 |
| Canada [C0006823] | 29 | 1 | 39 | 30 |
| Hippocampus [C0019564] | 26 | 73 | ||
These results indicate that the instances of California and Canada were distinguishable from each other but not Calcium and Hippocampus. Therefore, if the algorithm identified the target word as being a geographical location, the algorithm disambiguated the acronym correctly. We believe this is because the terms in the instances for the geographical locations were distinct. For example, terms such as alberta, inuit and saskatchewan existed in instances where CA referred to Canada but not California, and similarly, silicone, sun and copper exist in instances referring to California but not Canada.
The analysis of ca also shows that when the algorithm did not identify an instance as a geographical location, it randomly assigned the instance either Calcium or Hippocampus. We believe this is because approximately half of the mapped terms in instances referring to one sense also existed in instance referring to the other sense. For example, 203 out of 323 terms in instances referring to Hippocampus were also in instances referring to Calcium. Analysis of the target words hemlock, heregulin, and tomography showed similar results.
This was not the case for lawsonia. The target word lawsonia has two possible senses: Lawsonia [C1068388] the plant genus of the family Lythraceae that is the source of henna, and Lawsonia [C0752045] the genus of a bacteria. The confusion matrix in Table 5 shows that the possible senses look randomly assigned to the instances. Analysis of the terms in the instances though show that only 23 out of the 241 terms existing in instances referring to lawsonia the plant also existed in instances referring to the bacteria indicating that the context should have been distinct enough to disambiguate between the terms. This is verified when looking at the results of the path-based measure wup which obtained an overall disambiguation accuracy of 0.87 for this target word.
Table 5:
Error Analysis of the Target Word Lawsonia using Lin with a Window Size of 50
| Lawsonia Plant Genus C0752045 | Lawsonia Bacteria Genus C1068388 | |
|---|---|---|
| Lawsonia Plant Genus [C0752045] | 41 | 58 |
| Lawsonia Bacteria Genus [C1068388] | 9 | 7 |
As previously noted, lin and wup are similar, differing only in that wup uses the depth of the concept while lin uses the IC of the concept in the similarity calculation. Analysis of the possible senses of lawsonia shows that the depth of the concepts differ, the maximum depth is 9 for lawsonia the plant (C0752045) and 11 for lawsonia the bacteria (C1068388), but the IC for both of the senses are equal (5.79). This in effect removes the denominator from the equation in the lin measure and the difference in similarity is based only on the IC of the LCS which is equal to the similarity measure proposed by Resnik (res) multiplied by two. In the case of the UMLS::SenseRelate algorithm, the results obtained by res and lin would be the same, and as expected the results show that the overall disambiguation accuracy obtained by res is equal to that of lin for the target word lawsonia (0.43). Therefore, in the case where the possible senses have the same IC, the lin measure backs off to the res measure.
Further analysis of the individual target word results shows that there exist 42 target words (including lawsonia) whose possible senses have the same IC score. Of those target words, only eight obtain an overall disambiguation accuracy lower when using wup than when using lin. Of those eight, only two obtain an accuracy greater than 10 percentage points. This coincides with our previous finding that, although res obtains a statistically significantly lower overall disambiguating accuracy than lin, it is statistically significantly higher than any of the path-based measures including wup.
With respect to using various window sizes, the results in Table 1 show that words within a window size of 50 of the target word obtain the highest disambiguation accuracy. After 50, the accuracy remains the same or degrades. Not every term in the window of context mapped to a concept in MSH. Table 6 shows the number of concepts used by the IC measures and the path-based measures for the various window sizes. A window size of zero results in no terms or words being used which essentially resorts to a random assignment of the senses. These results show for a window size of 50 approximately 13 terms mapped to concepts. This indicates that locally occurring terms provide a sufficient enough of a distinction to determine of the sense of the target word. This is consistent with the finding reported by Choueka and Lusignan23 who conducted an experiment to determine what size window is needed for humans to determine the appropriate sense of an ambiguous word.
Table 6:
Number Terms Mapping to Concepts based on Window Size
| window size | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| measures | 0 | 1 | 2 | 5 | 10 | 25 | 50 | 60 | 70 |
| path-based | 0 | 0.27 | 0.83 | 1.97 | 3.72 | 8.16 | 13.67 | 14.28 | 16.86 |
| IC-based | 0 | 0.25 | 0.79 | 1.85 | 3.49 | 7.60 | 12.96 | 14.28 | 15.64 |
The results also show that the number of mappings is slightly higher for the path-based measures than the IC-based measures. This is because not all concepts have an information content and therefore the similarity can not be obtained. For example, the concept for drug induced liver injury (C2717837) was not found in our corpus and has an information content of zero.
Conclusions
In this paper, we evaluated a novel knowledge-based method for WSD, called UMLS::SenseRelate, that does not require manual annotation and yields a disambiguation accuracy sufficiently high for most practical purposes.
The objective of this work was to evaluate a method that can disambiguate terms in biomedical text using similarity information extrapolated from the UMLS, and evaluate the efficacy of IC-based semantic similarity measures. To do this, we evaluated UMLS::SenseRelate on the various semantic similarity measures in UMLS::Similarity and found that IC-based measures obtain a statistically significantly higher overall disambiguation accuracy than path-based measures. We believe this is because the IC-based measures weight the path based on where it exists in the taxonomy using the probability of the concepts occurring in a corpus of text.
Our study constitutes a significant step forward in the area of word sense disambiguation, as it will enable the incorporation of a scalable term disambiguation application into NLP systems used for indexing and retrieval of documents in the biomedical domain. It also provides a platform in which measures of semantic similarity and relatedness can be evaluated.
Future Work
In this work, we evaluated our method on the task of target-word disambiguation in which an instances containing a single target word are given to the system for disambiguation. In the future, we plan to extend the method in order to perform all-words disambiguation which disambiguates terms in a running text. In this process, we plan to incorporate the concept mapping system MetaMap24. Currently, the terms are obtained from the SPECIALIST Lexicon and are mapped to concepts using a dictionary look up, we plan to use MetaMap to identify the terms surrounding the target word and their mappings to the UMLS. The possible senses of a target word come from two sources, either directly from the UMLS using the MRCONSO table or a predefined set. In the future, we plan to use MetaMap to determine the possible senses of a target word.
We also plan to explore different ways at determining the window size in which to obtain context information and various ways to control the size of the window, for example, rather than the window containing terms that might or might not map to concepts in the UMLS, we plan to explore having the window contain only concepts.
Additionally, in our analysis of UMLS::SenseRelate, we found that using locally occurring terms obtains a higher disambiguation accuracy. In the future, we are considering weighting the terms based on their distance from the target word.
Furthermore, in this study the path information for the similarity measures was obtained from MSH. In the future, we plan to evaluate the effect of using different combinations of sources in order to determine their benefits and disadvantages. This would also allow us to evaluate UMLS::SenseRelate on datasets whose possible senses come from multiple sources and compare our method directly to previously proposed methods such as those discussed in the Related Work section.
Acknowledgments
This work was supported by the Grant #R01LM009623-01 from the National Institute of Health, National Library of Medicine.
We would like to thank Russel Loane, Jim Mork and Lan Aronson from the National Library of Medicine for providing the UMLSonMedline dataset.
Footnotes
A complete listing of the individuals results for all the terms can be downloaded at http://rxinformatics.umn.edu/
References
- 1.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation. Journal of the American Medical Informatics Association. 2004;11(4):320–331. doi: 10.1197/jamia.M1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leroy G, Rindflesch TC. Effects of information and machine learning algorithms on word sense disambiguation with small datasets. International Journal of Medical Informatics. 2005;74(7–8):573–85. doi: 10.1016/j.ijmedinf.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Joshi M, Pedersen T, Maclin R. A comparative study of support vectors machines applied to the supervised word sense disambiguation problem in the medical domain. Proceedings of 2nd Indian International Conference on Artificial Intelligence; Dec, 2005. pp. 3449–3468. [Google Scholar]
- 4.McInnes B, Pedersen T, Carlis J. Using umls concept unique identifiers (cuis) for word sense disambiguation in the biomedical domain. Proceedings of the Annual Symposium of the American Medical Informatics Association; Nov, 2007. [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson M, Guo Y, Gaizauskas R, Martinez D. Disambiguation of biomedical text using diverse sources of information. BMC Bioinformatics. 2008;911(Suppl 11) doi: 10.1186/1471-2105-9-S11-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan JW, Friedman C. Word Sense Disambiguation via Semantic Type Classification. Proceedings of the American Medical Informatics Association Symposium; Nov, 2008. pp. 177–181. [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen T. The Effect of Different Context Representations on Word Sense Discrimination in Biomedical Texts. Proceedings of the 1st ACM International Health Informatics Symposium; Nov, 2010. pp. 56–65. [Google Scholar]
- 8.Humphrey SM, Rogers WJ, Kilicoglu H, Demner-Fushman D, Rindflesch TC. Word sense disambiguation by selecting the best semantic type based on journal descriptor indexing: Preliminary experiment. Journal of the American Society for Information Science and Technolology. 2006;57(1):96–113. doi: 10.1002/asi.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexopoulou D, Andreopoulos B, Dietze H, Doms A, Gandon F, Hakenberg J, Khelif K, Schroeder M, Wachter T. Biomedical word sense disambiguation with ontologies and metadata: automation meets accuracy. BMC Bioinformatics. 2009;10(1):28. doi: 10.1186/1471-2105-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rada R, Mili H, Bicknell E, Blettner M. Development and application of a metric on semantic nets. IEEE Transactions on Systems, Man, and Cybernetics. 1989;19(1):17–30. [Google Scholar]
- 11.Caviedes JE, Cimino JJ. Towards the development of a conceptual distance metric for the umls. Journal of Biomedical Informatics. 2004;37(2):77–85. doi: 10.1016/j.jbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, Palmer M. Verbs semantics and lexical selection. Proceedings of the 32nd Meeting of Association of Computational Linguistics; 1994. pp. 133–138. [Google Scholar]
- 13.Leacock C, Chodorow M. Combining local context and WordNet similarity for word sense identification. WordNet: An electronic lexical database. 1998;49(2):265–283. [Google Scholar]
- 14.Nguyen HA, Al-Mubaid H. New ontology-based semantic similarity measure for the biomedical domain. Proceedings of the IEEE International Conference on Granular Computing; 2006. pp. 623–628. [Google Scholar]
- 15.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. Proceedings of the 14th International Joint Conference on Artificial Intelligence; 1995. pp. 448–453. [Google Scholar]
- 16.Jiang J, Conrath D. Semantic similarity based on corpus statistics and lexical taxonomy. Proceedings on International Conference on Research in Computational Linguistics; 1997. pp. 19–33. [Google Scholar]
- 17.Lin D. An information-theoretic definition of similarity. Proceedings of the International Conference on Machine Learning; 1998. pp. 296–304. [Google Scholar]
- 18.McInnes BT, Pedersen T, Pakhomov SV. UMLS-Interface and UMLS-Similarity : Open Source Software for Measuring Paths and Semantic Similarity. Proceedings of the American Medical Informatics Association Symposium; San Fransico, CA. November 2009; [PMC free article] [PubMed] [Google Scholar]
- 19.Patwardhan S, Banerjee S, Pedersen T. Using measures of semantic relatedness for word sense disambiguation. Proceedings of the Fourth International Conference on Intelligent Text Processing and Computational Linguistics; 2003. pp. 241–257. [Google Scholar]
- 20.Burgun A, Bodenreider O. Comparing terms, concepts and semantic classes in WordNet and the Unified Medical Language System. Proceedings of the NAACL Workshop on WordNet and Other Lexical Resources: Applications, Extensions and Customizations; 2001. pp. 77–82. [Google Scholar]
- 21.Ide NC, Loane RF, Demner-Fushman D. Essie: a concept-based search engine for structured biomedical text. Journal of the American Medical Informatics Association. 2007;14(3):253–263. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimeno-Yepes A, McInnes BT, Aronson AR. Exploiting MeSH indexing in MEDLINE to generate a data set for word sense disambiguation(accepted) To Appear in BMC bioinformatics. 2011 doi: 10.1186/1471-2105-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choueka Y, Lusignan S. Disambiguation by short contexts. Computers and the Humanities. 1985;19(3):147–157. [Google Scholar]
- 24.Aronson A. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. Proceedings of the American Medical Informatics Association Symposium; 17–21, November 2001; [PMC free article] [PubMed] [Google Scholar]