Figure 5.
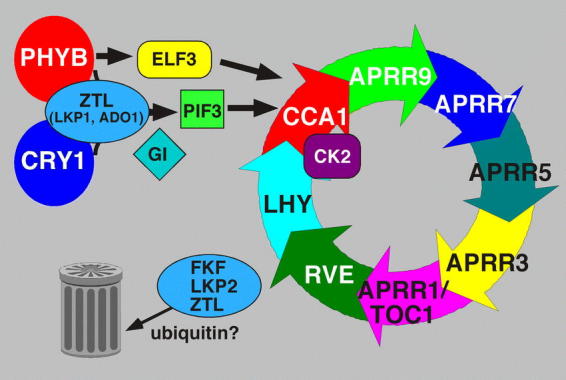
A speculative model of an Arabidopsis circadian clock. Light input via phytochromes and cryptochromes (for simplicity, only PHYB and CRY1 are shown, although others are certainly involved) is mediated through ZTL, ELF3 and GI, or through PIF3. ZTL/ADO1 is known to bind to PHYB and CRY1. PIF3 binds to CCA1 and LHY promoters and possibly to other targets in the clock. The pathway downstream of GI is not known. Although the input pathways are drawn as discrete linear pathways, there may be interaction among them. For simplicity, a single central oscillator is illustrated with a number of putative oscillator components indicated. Components on the circular arrows oscillate in mRNA or protein abundance. One should not infer causal relationships among putative components from the relative order of their placement on the circle as experimental proof is lacking. FKF/LKP2/ZTL are clustered, although there is no evidence that they form molecular complexes. Moreover, LKP2 mRNA oscillates and overexpression results in arrhythmicity, making LKP2 a strong candidate as an oscillator component. CCA1 and LHY are phosphorylated by CK2, which may make them substrates for the F-box proteins (ZTL, FKF and LKP2) and target them for ubiquitination and degradation by the proteasome (trash can). Output pathways may emanate from any of the putative oscillator components. CCA1, LHY, RVEs and TOC1/APRR1 are DNA-binding proteins, and CCA1 is know to bind to LHCB promoters. Other outputs from the oscillator feed back to input components, such as PHYA, PHYB and CRY1, which are regulated by the clock at transcriptional and mRNA abundance levels
