Figure 4.
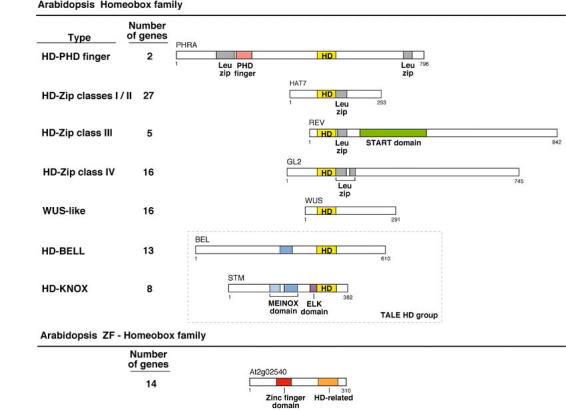
The Arabidopsis Homebox (HB) and ZF-HB gene families. The Arabidopsis homeobox gene family can be subdivided into different groups according to the combinations of domains that the corresponding proteins contain, and to the phylogenetic analysis of the homeodomain. The number of members in each Arabidopsis homeobox gene subfamily is indicated (except for three genes, whose classification is unclear and that are not represented in this figure). Most of the combinations of a homeodomain with a domain of a different type (leucine zipper, PHD finger, START domain) are the result of domain shuffling events specific to the plant kingdom: those combinations are not found in Drosophila, C. elegans, or yeast homeodomain proteins. The only Arabidopsis homeodomain proteins that have an additional motif also found in animal homeodomain proteins are those of the KNOX class, which contain a MEINOX domain (Bürglin, 1998). Conversely, homeodomains in animals are associated with a large variety of motifs, such as the paired and POU-specific domains (which are themselves specific to animals), the LIM motif, or C2H2 zinc fingers, in combinations that are not present in Arabidopsis (for a similar depiction of the animal homeodomain proteins, see Gehring et al., 1994). The START domain is a lipid-binding domain that could provide regulation of HD-Zip class III protein function by sterols. It is found in a variety of eukaryotic proteins, but has been found associated with transcription factor domains only in this class of plant homeobox genes (Ponting and Aravind, 1999). Proteins of the ZF-HB family contain a homeodomain-related sequence that is more divergent from all the different groups of homeodomain sequences of the HB family than these are among themselves (Windhövel et al., 2001). In addition, ZF-HB proteins contain a plant-specific zinc coordinating motif (Windhövel et al., 2001)
