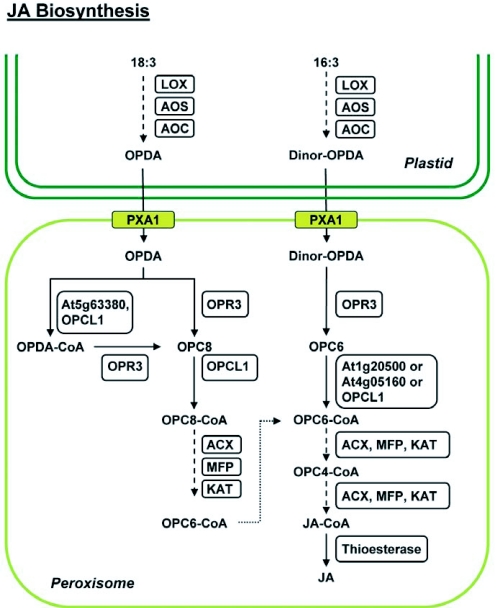
Figure 5.
JA biosynthesis pathway.
Biosynthesis of (+)-7-iso-jasmonic acid (JA) initiates in the chloroplast with the sequential action of lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC) on linolenic acid (C18:3) or hexadeca-trienoic acid (C16:3) to generate 12-oxophytodienoic acid (OPDA) and dinor-OPDA, respectively. Transport of OPDA and dinor-OPDA into peroxisomes is facilitated either by the ABC transporter (PXA1) or by an uncharacterized pathway (not shown). Oxophytodienoic acid reductase 3 (OPR3) reduces OPDA to 3-oxo-2-(2′-[Z]-pentenyl) cyclopentane-1-octanoic acid (OPC8) and dnOPDA to 3-oxo-2-(2′-pentenyl)-cyclopentane-1-hexanoic acid (OPC6), with the resultant compounds being activated to their corresponding CoA esters by OPC:8 CoA ligasel (OPCL1) or the indicated acyl-CoA synthetases (At1g20500, At4g05160, At5g63380). The CoA derivatives undergo β-oxidation by the consecutive activities of acyl-CoA oxidase (ACX), multifunctional protein (MFP) and 3-ketoacyl-CoA thiolase (KAT), eventually generating JA-CoA after being subjected to the requisite number of β-oxidation cycles (3 for OPCS-CoA and 2 for OPC6-CoA). A putative thioesterase cleaves the CoA moiety, releasing JA. Dotted arrows depict catalysis of substrate(s) by the consecutive actions of the indicated enzymes.