Figure 3.
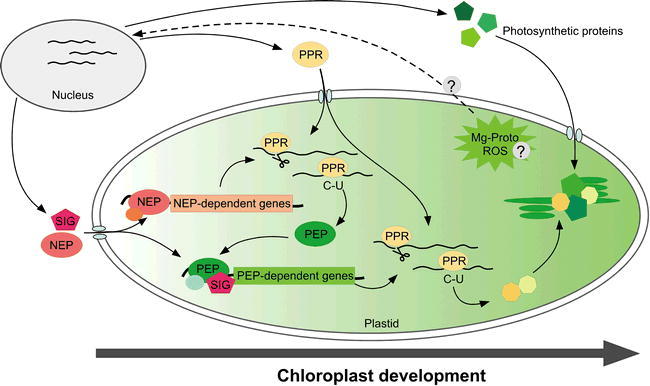
A regulatory network of nuclear and chloroplast gene expression.
This schematic view represents chloroplast gene expression and the assembly of photosynthetic proteins. The process is governed by the coordinated transcription mediated by the NEP and PEP polymerases, and by the post-transcriptional regulatory steps mediated by PPR proteins. A time-course of chloroplast development is illustrated spatially, from left to right. The flow of gene products (NEP, SIG, PPR, and photosynthetic proteins) is indicated by arrows. At an initial stage, NEP and SIG are synthesized and imported into proplastids. These molecules drive the subsequent expression of NEP-dependent genes, including PEP, and lead to the ‘switching-on’ of chloroplast transcription. Numerous PPR proteins are concomitantly imported from the cytosol, and play roles in RNA processing, editing and translation. The products of photosynthetic genes in the chloroplast genome are finally assembled into complexes with other subunits encoded by the nuclear genome, the latter components having been synthesized in the cytosol and imported. To enable coordinated regulation between the nuclear and chloroplast genomes, Mg-protoporphyrin-IX (Mg-proto) and ROS act as possible retrograde signals (indicated by the dotted line); the precise nature of these retrograde signaling pathways is not clear at the present time.
