Background: IgG that protects children from meningococcal disease binds lipooligosaccharides, but the antigen was not known.
Results: The lacto-N-neotetraose α chain conforms at least four antigens; IgG that killed strains with a lacto-N-neotetraose α chain bound internal antigens.
Conclusion: Only the internal lacto-N-neotetraose antigens induce broadly protective IgG.
Significance: Induction of IgG that recognizes internal lacto-N-neotetraose antigens by vaccination would prevent endemic meningococcal disease.
Keywords: Antibodies, Glycobiology, Immunochemistry, Infectious Diseases, Mass Spectrometry (MS), Oligosaccharide, Vaccine Development, Neisseria meningitidis, Lipooligosaccharides
Abstract
Antibodies that initiate complement-mediated killing of Neisseria meningitidis as they enter the bloodstream from the oropharynx protect against disseminated disease. Human IgGs that bind the neisserial L7 lipooligosaccharide (LOS) are bactericidal for L3,7 and L2,4 meningococci in the presence of human complement. These strains share a lacto-N-neotetraose (nLc4) LOS α chain. We used a set of mutants that have successive saccharide deletions from the nLc4 α chain to characterize further the binding and bactericidal activity of nLc4 LOS IgG. We found that the nLc4 α chain conforms at least four different antigens. We separately purified IgG that required the nLc4 (non-reducing) terminal galactose (Gal) for binding and IgG that bound the truncated nLc3 α chain that lacks this Gal residue. IgG that bound the internal nLc3 α chain killed both L3,7 and L2,4 strains, whereas IgG that required the nLc4 terminal Gal residue for binding killed L2,4 stains but not L3,7 strains. These results show that the diversity of LOS antibodies in human serum is as much a function of the conformation of multiple antigens by a single glycoform as of the production of multiple glycoforms. Differences in sensitivity to killing by human nLc4 LOS IgG may account for the fact that fully two-thirds of endemic group B meningococcal disease in infants and children is caused by L3,7 strains, but only 20% is caused by L2,4 stains.
Introduction
Neisseria meningitidis is a unique commensal of the human pharynx that occasionally causes sepsis and meningitis that can be fulminate and rapidly fatal (1). The asymptomatic commensal relationship is maintained by circulating antibodies that initiate complement-mediated killing of the organisms as they enter the bloodstream from the pharynx (2–4). Bactericidal antibodies in the newborn's serum are IgGs of maternal origin (2), but they are rapidly catabolized and only slowly and irregularly replaced by antibodies induced, first, by colonization of the pharynx by Neisseria lactamica, a related bacterium that can split lactose, the disaccharide in milk (5), and then by unencapsulated and therefore essentially avirulent strains of N. meningitidis (5–7). By age 10, 45–75% of children have regained serum bactericidal activity (2). The window of susceptibility between the loss of maternal IgG and the autonomous acquisition of bactericidal antibodies correlates with the peak incidence of meningococcal disease (2). The antigenic specificity of protective antibodies has long been sought (8), because their induction by vaccination in infancy would be the most straightforward approach to the prevention of endemic disease.
N. lactamica share lipooligosaccharide (LOS)3 epitopes with N. meningitidis but lack common capsular and class 1, 2, and 3 protein epitopes (9). This suggests that the earliest bactericidal antibodies induced in infants may be LOS-specific. However, because each LOS glycoform molecule conforms multiple different epitopes (10, 11), and each meningococcal strain makes several different LOS glycoforms (12), it has been difficult to sort out just which LOS antigens induce protective antibodies (10, 11, 13).
We recently reported a means of affinity-purifying human IgG that binds different LOS structures and showed that these antibodies can kill group B endemic case isolates in the presence of human complement (14). One IgG pool bound LOS made by the Neisseria gonorrhoeae strain, 1291, which have the paraglobosyl lacto-N-neotetraose (nLc4)4 structure as their α chain (15). These IgGs were bactericidal for meningococci that have nLc4 α chains (16–18), as adduced by their LOS immunotypes (19), and that account for ∼85% of endemic meningococcal cases (12). N. lactamica also make LOS with the nLc4 α chain (9).
Those data supported the hypothesis that LOS antibodies contribute to natural immunity, but because the strains were characterized only by LOS immunotype, which incompletely captures LOS diversity (12, 21, 22), the exact LOS structures to which the antibodies bind could not be determined. Such detailed information is needed before the question of how best to induce protective LOS IgG in infants can be addressed.
In order to define the specificity of LOS IgG and the breadth of protection that they might afford, we used LOS from pyocin C-selected mutants of strain 1291 that have successive saccharide deletions from the nLc4 α chain to affinity-purify LOS IgG (14, 15). We then performed genetic analyses of endemic group B case strains and mass spectrometry of intact LOS molecules to provide structural information beyond immunotype. We report that the full-length nLc4 LOS presents at least four antigens. Our data show that IgG that requires the nLc4 terminal galactose (Gal) for binding is not bactericidal for L3,7 strains that account for roughly two-thirds of endemic cases but readily kills L2,4 strains that account for about 20% of endemic disease (10–13). IgG that efficiently kills L3,7 strains in the presence of human complement bind one or more internal antigens within the truncated nLc3 glycolipid. These antibodies also efficiently kill L2,4 strains. Their induction during infancy would protect infants and young children from 85% of endemic group B meningococcal disease.
EXPERIMENTAL PROCEDURES
Bacteria
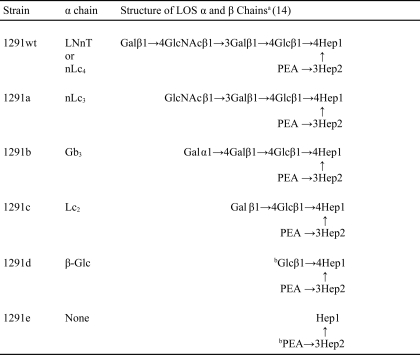
Strains are described in Tables 1 and 2. N. gonorrhoeae strain 1291 is the Gc2 prototype strain in the Apicella serogrouping system (23). Its LOS has an nLc4 α chain that lacks the terminal N-acetylgalactosamine (GalNAc) substitution common to most gonococcal LOS (24, 25), no β chain, and an O-acetylated, N-acetylglucosamine (GlcNAc) γ chain (15). There is a single phosphoethanolamine (PEA) substitution of the second heptose (Hep2) at C3 (15). This is the same structure as that of the L7 meningococcal LOS (16). The 1291 mutants are denominated 1291a–e, based on the electrophoretic mobility of their LOS through SDS-PAGE (26). For this study, we used LOS made by 1291wt and 1291a to affinity-purify LOS IgG.
TABLE 1.
LOS of N. gonorrhoeae 1291 and its mutants
a Gal, galactose; GlcNAc, N-acetylglucosamine; Glc, glucose; Hep, heptose; PEA, phosphoethanolamine.
b 1291d and 1291e each can make various amounts of both of the LOS glycoforms shown.
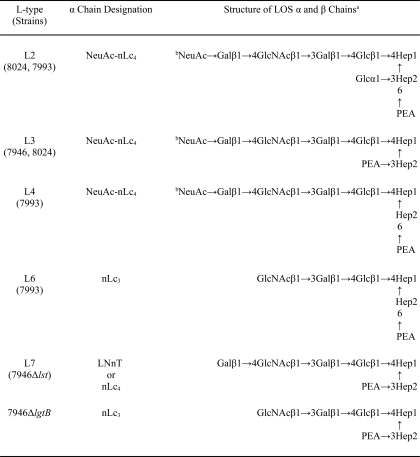
TABLE 2.
a NeuAc, N-acetylneuraminic (sialic) acid; Gal, galactose; GlcNAc, N-acetylglucosamine; Glc, glucose; Hep, heptose; PEA, phosphoethanolamine.
b Only a portion of the nLc4 glycoforms are sialylated.
We used the SPRIA inhibition technique of Zollinger and Mandrell (19), with their original polyclonal rabbit antisera, to determine the LOS immunotype(s) (L-types) of 34 consecutive and unique N. meningitidis isolates from the blood or cerebrospinal fluid of epidemiologically unrelated infants and children hospitalized during a 15-month period of endemic disease in Houston, Texas (10–13) and selected three group B strains, 7946 (L3,7), 8024 (L2,3), and 7993 (L2,4,6,7), for this study. The Zollinger and Mandrell system arbitrarily sets 35% inhibition by a test strain of the binding of a type-specific antiserum to its homologous prototype strain as the threshold criterion for a serotype designation; inhibition of prototype-antiserum pairs by <35% is ignored (19). Structures assigned to the serotypes of the strains (16–18, 20) are in Table 2. All organisms were cultured as described previously (12–14).
Sequencing the 1291 lgt Operon
Although the structures of LOS made by 1291wt and its pyocin-resistant mutants were known (15), the only known mutations were those of 1291d and 1291e, which both have mutations in pgm, which encodes the neisserial phosphoglucomutase (27). We used PCR to amplify lgtABCDE from 1291wt and the 1291a–c mutants with the use of primers in Table 3 (28, 29) and sequenced the resulting amplicons. We used the DNA sequence numbering for the lgtABCDE region of gonococcal strain, F62, as described in the National Center for Biotechnology Information database under accession number U14554 to orient the sequences to the literature (29).
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequence | Source or reference |
|---|---|---|
| CB-5 | GCCGGCATCGAGGACGTGGAACCTGA | Ref. 28 |
| JL-12 | AGCGGCCCATCCCGATACGGA | Ref. 30 |
| JL-50 | GGCCGACATCGCGCTTTTGGGCG | Ref. 29 |
| JL-51 | GGGGCGATTTTACCTAGCAGATGAA | Ref. 29 |
| LgtB F | GCTTCCGCCGCAGAACGCAG | This study |
| LgtB R | CTGTTCGCGCCTTTGCCGGC | This study |
| Lst F | CGGCATATTTTCGGTGAAAT | This study |
| Lst R | CCTTATTTTTACCCCACGCA | This study |
| KanF | TTTATGGACAGCAAGCGAAC | This study |
| KanR | GGAGAAAATACCGCATCAGG | This study |
| lpt3 F | CGCTGACATTTGTGATTGCT | This study |
| lpt3 R | TATCGTAGCCCAACGTGTGA | This study |
| lpt6 F | ATTTTTGTTTGTAACGGCGG | This study |
| lpt6 R | GTCTTTGGCTTCCAGACCAG | This study |
| lgtGin F | ACGCTCATTCCTCTTTTCAA | This study |
| lgtGin R | CCATAATGGTAAAGTCGGCG | This study |
| 6GF F | GAAACCCTGTCCAAATTC | This study |
| 6GF R | GAACCAGATGCCGTTGATTT | This study |
PCR mixtures contained 2 μm primer pairs (Integrated DNA Technologies, Coralville, IA), 1 mm dNTPs (Invitrogen), 1% DMSO (Finzymes, Espoo, Finland), 1× GC buffer (Finzymes), and 0.5 ml of Phusion High Fidelity Polymerase (Finzymes). The PCR program used was as follows: denaturation at 99 °C for 10 min, followed by 10 cycles of 30 s at 98 °C, 30 s at 64 °C, and 5 min 20 s at 72 °C and then 30 cycles during which the annealing temperature was 68 °C. Final extension was for 7 min at 72 °C.
Amplicons were isolated from a preparative 1.4% agarose gel with use of the Qiagen gel extraction kit (Mountain View, CA), according to the manufacturer's instructions, and then cloned into the pCR4-TOPO vector using the TOPO TA Cloning Kit for Sequencing (Invitrogen). One Shot TOP10 chemically competent cells provided with the kit were used to generate transformants, which were plated onto Luria-Bertani plates that contained 50 μg/ml kanamycin. Plasmid minipreps of transformants were analyzed by restriction digestion with EcoRI (New England Biolabs, Ipswich, MA). Digestion products were visualized in 1.4% agarose gels with SBRY Safe stain (Invitrogen). Plasmids from transformants with correct insert size were purified using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer's instructions and sequenced by Sequetech (Valencia, CA).
LOS
The late Herman Schneider (Walter Reed Army Institute of Research) purified LOS from 1291wt and 1291a (30). LOS were purified from other strains with the use of the same modified hot phenol/water method (30, 31).
Genotyping N. meningitidis LOS
We determined the LOS genotypes of 7946, 8024, and 7993 by sequencing lgtA, lgtB, lgtE (28, 32), lgtG (33), lpt3 (34), and lpt6 (35) with the use of primers in Table 3. The 6GF primers amplify both lpt6 and lgtG, which are on opposite strands, along with short flanking regions. lgtGin amplifies an internal lgtG segment; lpt6 amplifies lpt6.
Construction of LOS Mutants
Group B N. meningitidis endogenously sialylate a portion of their LOS molecules (12, 36–37). LOS sialylation is catalyzed by an α-2,3-sialyltransferase (Lst) (38). We inactivated lst (38) and lgtB (28) in 7946 by insertion of antibiotic resistance cassettes to generate mutant strains, 7946Δlst and 7946ΔlgtB. lst and lgtB were amplified by PCR with the use of primers in Table 3. The PCR products were cloned into EcoRI restriction sites of pUC19. The kanamycin resistance gene was amplified by PCR (Table 3) and inserted into the AgeI site of lst or the BssHII site of lgtB. The plasmids carrying the inactivated genes then were transformed into 7946. Transformants were selected on GC agar plates that contained 50 μg/ml kanamycin, and the mutants were confirmed by PCR, SDS-PAGE of proteinase K (PK) lysates (30, 39), and mass spectrometry of O-deacylated LOS (14, 40).
SDS-PAGE
We screened LOS made by the mutants by SDS-PAGE (30) of PK-treated whole cell lysates from overnight growth (39) with use of a Mini-Protean 3 cell (Bio-Rad) (14). For optimal resolution, LOS were electrophoresed through 27 cm of a 13.1% polyacrylamide gel with an SE600 series vertical slab gel unit (Hoefer, San Francisco, CA) (41). LOS were visualized with silver stain (12, 30).
Mass Spectrometry
Intact 7946 and 8024 LOS and O-deacylated LOS of 7946 and its Δlst and ΔlgtB mutants were analyzed by negative ion matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), as described (40, 42, 43). MALDI-TOF MS was performed in the linear as well as the reflectron mode on a Voyager-DE STR instrument equipped with a 337-nm nitrogen laser and delayed extraction. Spectra were processed with digital smoothing and base-line correction using Data Explorer software. The spectra of the intact LOS were externally calibrated using the average fragment and molecular ion masses of the intact 1291wt LOS at m/z 1836.3, m/z 1881.6, and m/z 3718.9; a one-point internal calibration using the mass of the lipid A fragment ions at m/z 1836.3 also was used for the 8024 spectrum. The spectra of the O-deacylated 7946, 7946Δlst, and 7946ΔlgtB LOS were acquired and externally calibrated using ions for intact 7889 LOS at m/z 1836.3 and m/z 3676.6 or 89I LOS at m/z 1738.3 and m/z 4090.1 (42, 43), and then internally calibrated using either one or two calculated masses for peaks at m/z 952.0, m/z 1677.4, m/z 1839.9, and m/z 2792.6. Structures were assigned to molecular ions on the basis of composition, previously published structures (12, 15–18, 20, 42–44), L-type (19), and the genetics of LOS synthesis (45).
Affinity Purification of Human LOS Antibodies
Human LOS IgGs were isolated as described (14). Briefly, purified LOS from strains 1291wt and 1291a were disaggregated in 3% deoxycholic acid (46) in 0.2 m carbonate buffer (pH 10.2) and then coupled to epoxy-activated Sepharose 6B (Amersham Biosciences) through the epoxy linkage. Immune Globulin Intravenous (Human) (IGIV) (CSL Behring AG, Bern, Switzerland) was used as the antibody source. 1 g diluted in 10 ml of phosphate-buffered saline (PBS) was loaded onto a column and allowed to bind to LOS. After washing, the bound IgG was eluted with glycine-HCl (pH 2.3), and the pH of the eluate was immediately adjusted to pH 7–8 with 1 m Tris, followed by dialysis against PBS.
To isolate IgG that bound nLc4 LOS, IGIV was passed through the 1291wt column, and IgG was eluted from the column. To isolate IgG that bound nLc3 LOS, IGIV was passed through the 1291a column, and IgG was eluted. To isolate IgG that required the nLc4 terminal Gal for binding, IGIV was passed through the 1291a column multiple times to ensure complete removal of nLc3 IgG and then passed through the 1291wt gel, after which the IgG was eluted; this IgG is denominated nLc4-nLc3 IgG.
Whole-cell ELISA
A whole-cell ELISA (47) was used to analyze binding of IgG. Briefly, organisms were harvested from overnight growth on GC agar plates, washed, and suspended in PBS to an optical density (OD) at 640 nm of 0.1. The suspension was heated at 56 °C for 1 h and kept at 4 °C overnight. 100 μl of the suspension was used to coat microtiter wells, which then were blocked with 1× Roche Applied Science blocker, washed with PBS, and incubated with either 10 μg/ml or serial dilutions of IgG. After washing, goat anti-human IgG conjugated with alkaline phosphatase (Sigma-Aldrich) was added into each well as secondary antibody. After incubation, the wells were washed and developed with 4-nitrophenyl phosphate (Sigma-Aldrich). The absorbance at 405 nm was determined with a Thermomax microplate reader (Molecular Devices, Sunnyvale, CA). Negative control wells received everything except IgG.
Bactericidal Activity
The bactericidal assay has been described (14). Briefly, bacteria were harvested after overnight growth on GC agar and grown to mid-log phase in GC broth with 1% IsoVitaleX. Bacteria were washed, suspended in gonococcal buffer, and adjusted to an OD at 580 nm of 0.1 (105 cell/ml). The reaction mixture contained 104 organisms/ml, 10–40% NHS, and various concentrations of human IgG. Control reactions contain only NHS or heated inactivated NHS. The tubes were incubated with end-over-end rotation at 37 °C. At time 0 and 60 min, duplicate 10-μl aliquots were removed and plated. Colony-forming units (cfu) were determined after overnight growth. Survival was expressed as the percentage of cfu present at time 0 that survived after 60 min.
NHS was obtained from one of the authors (J. M. G.) who has no history of neisserial infection or meningococcal carriage, and the concentration was adjusted to maximize the complement concentration while minimizing any intrinsic bactericidal activity against each strain. Each assay was repeated in duplicate at least twice.
Statistical Analysis
Statistical analyses of the binding of IgG to the 1291 mutants were performed using SigmaStat for Windows version 3.11 (Systat Software, San Jose, CA). When ANOVA found significant differences among multiple measurements, groups of data were analyzed by the Student-Newman-Keuls method for multiple pairwise comparisons. Analyses of paired data were done using the Excel Student's t test program. Values of p < 0.05 were considered significant for all comparisons.
RESULTS
1291 LOS
1291wt has a +2 frameshift in its lgtD poly(G) tract that results in early termination of lgtD transcription and that accounts for the absence of a terminal α chain GalNAc residue. 1291a has a 2867-bp deletion from bp 213 of lgtB to bp 223 of lgtE that results in deletion of lgtC and lgtD and translation of an LgtB/E fusion protein.
1291b has a +1 frameshift in its lgtA poly(G) tract (15 G residues) that results in early termination of lgtA transcription and that accounts for the absence of the terminal N-acetyllactosamine of the nLc4 α chain. Instead, it has the LgtC-catalyzed globosyl Gb3 α chain, which is not made by the parent strain.
1291c has a +2 frameshift in its lgtA poly(G) tract (13 G residues) and a 2875-bp deletion from the end of the lgtA poly(G) tract through the lgtD poly(G) tract that results in deletion of lgtB and lgtC and in early termination of translation of an LgtA/D fusion protein. These mutations are consistent with the structures in Table 1.
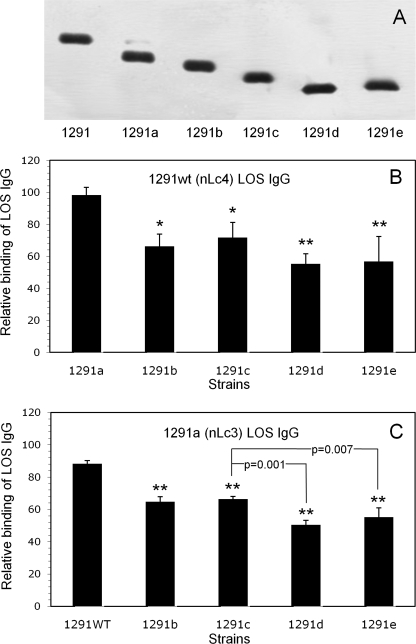
SDS-PAGE of the LOS of the 1291 mutants is shown in Fig. 1A; the LOS demonstrated the sequential loss of non-reducing terminal sugars shown in Table 1. 1291a made a very small amount of 1291wt LOS, and 1291e LOS was composed of two closely migrating species that together migrated more slowly than 1291d LOS. The second 1291e LOS, made in low abundance, would be the basal phosphoethanolaminyl diheptoside glycolipid plus the β-Glc substitution that initiates the α chain (Table 1 and Ref. 27).
FIGURE 1.
Binding of affinity-purified 1291wt (nLc4) LOS IgG and 1291a (nLc3) LOS IgG to 1291 and its pyocin-selected mutants. A, SDS-PAGE separated LOS from 1291 and each of the 1291 mutants. LOS were electrophoresed through 27 cm of 13.1% polyacrylamide gel and visualized with silver stain. The LOS α chain structures are in Table 1. Note that 1291a makes a small amount of 1291wt LOS and that 1291e LOS is composed of two closely migrating species that, together, migrate more slowly than 1291d. B, whole-cell ELISA of 1291wt (nLc4) LOS IgG. Microtiter wells were coated with the strains and then reacted with 10 μg/ml IgG. Binding is expressed as the percentage of the binding of the IgG to 1291wt (100%). Error bars, S.D. of three assays. Differences in binding to the strains were statistically significant (p = 0.002; ANOVA). Pairwise multiple comparisons (Student-Newman-Keuls method) revealed that the 1291b–e mutants bound significantly less nLc4 LOS IgG than 1291a (**, p = 0.002; *, p = 0.007). C, whole-cell ELISA of 1291a (nLc3) LOS IgG. Microtiter wells were coated with the strains and then reacted with 10 μg/ml IgG. Binding is expressed as a percentage of the binding of the IgG to 1291a (100%). Error bars, S.D. of three assays. Differences in binding to the strains were statistically significant (p < 0.001; ANOVA). Pairwise multiple comparisons revealed not only that the 1291b–e mutants bound significantly less nLc3 LOS IgG than 1291wt (**, p < 0.001) but that both 1291d (p = 0.001) and 1291e (p = 0.007) bound less than 1291c or 1291b. The difference in binding between 1291d and 1291e was not significant (p = 0.12).
Antigenic Structure of 1291 LOS
Fig. 1 also depicts the binding of 1291wt (nLc4) LOS IgG to each of the 1291 mutants, as compared with its binding to 1291wt, and as determined by whole-cell ELISA (Fig. 1B), and the binding of 1291a (nLc3) LOS IgG to 1291wt and each of the 1291b–e mutants, as determined by whole-cell ELISA and as compared with its binding to 1291a (Fig. 1C).
Binding of nLc4 LOS IgG to the mutants (Fig. 1B) was not uniform (p = 0.002; ANOVA); the 1291b–e mutants bound significantly less nLc4 LOS IgG than 1291a (p = 0.007 for 1291b and 1291c versus 1291a; p = 0.002 for 1291d and 1291e versus 1291a), which showed that the nLc4 LOS IgG was composed of more than one specificity. Because 1291wt does not make the 1291b LOS, IgG affinity-purified by its LOS would not contain 1291b specificity, and binding to this mutant would be to the internal 1291c structure that is proximal to the (non-reducing) terminal α-galactose of the 1291b Gb3 α chain.
Binding of nLc3 LOS IgG to 1291wt and the 1291b–e mutants (Fig. 1C) partially clarified the multiple specificities within nLc4 LOS IgG. Again, binding was not uniform (p = 0.001; ANOVA); not only did all four of the 1291b–e mutants bind significantly less nLc3 LOS IgG than 1291wt (p < 0.001), but both 1291d (p = 0.001) and 1291e (p = 0.007) bound less than 1291c or 1291b. Because 1291a does not make the 1291b LOS, IgG affinity-purified by its LOS would not contain 1291b specificity, and binding to 1291b would be to the internal 1291c structure. Thus, nLc3 LOS IgG was shown to contain at least three different specificities: itself, 1291c, and 1291d–e.
Binding of nLc3 LOS IgG to 1291wt was 12% less than to itself, which showed that 1291wt LOS conformed an antigen that was missing from 1291a. Because 1291a made a small amount of 1291wt LOS (Fig. 1A), the nLc4 LOS IgG bound 1291a as well as 1291wt, but the amount of 1291wt LOS made by 1291a did not appear to be sufficient to affinity-purify much 1291wt LOS IgG.
That the nLc4 LOS α chain conformed an antigen that was not conformed by the nLc3 α chain was confirmed by passage of IGIV first through the 1291a gel to remove nLc3 IgG and then through the 1291wt gel, followed by elution of nLc4 IgG. The yield of nLc4 LOS IgG from 1 g of IGIV was about 1185 μg, that of nLc3 LOS IgG was about 838 μg, and that of nLc4-nLc3 LOS IgG was about 206 μg. Thus, the yield of nLc4-nLc3 IgG from 1 g of IGIV was 17% of that of nLc4 IgG, a percentage reasonably close to the 12% estimated from the binding of nLc3 LOS IgG to 1291wt. Thus, nLc4 LOS IgG was shown to contain at least four different specificities: itself, 1291a, 1291c, and 1291d–e.
Taken together, these analyses enabled estimates of the four specificities within nLc4 LOS IgG. Roughly 15% of the IgG was specific for the nLc4 (non-reducing) terminal β-galactose, another 20% was specific for the 1291a nLc3 α chain, and 15% was specific for the 1291c Lc2 glycose. The remaining 50% of the IgG bound one or more of the antigens formed by the basal phosphoethanolaminyl diheptoside glycolipid and/or the γ chain GlcNAc. 1291e, which made a small amount of the β-Glc-substituted phosphoethanolaminyl diheptoside glycolipid, bound more of both IgG preparations than 1291d, which suggested that this basal glycose expressed an antigen not formed by the 1291d glycolipid, but the difference between the triplicate measurements was not significant (p = 0.12).
Antigenic Structures of Meningococcal LOS
The immunotype glycose structures are in Table 2; each strain made at least two different immunotypes. Based on SPRIA inhibition, ∼30–40% of the LOS molecules made by 8024 or 7993 have the L3 or L7 glycose, respectively. The L7 glycose is identical to that of 1291wt, and the L6 glycose is identical to that of 1291a, except for the location of the PEA substitution.
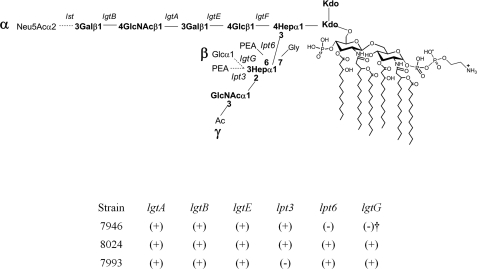
LOS Genotypes
The LOS genotypes were as expected from their L-types (Fig. 2). 7946 had an active lpt3, but lpt6 was missing, and lgtG was mutated and out of frame. 8024 and 7993 had active lpt6 and lgtG genes. 8024 also had an active lpt3, as would be needed for it to make the L3 glycoform, but lpt3 was missing from 7993.
FIGURE 2.
Schematic presentation of the biosynthetic genes responsible for N. meningitidis LOS structures and the genotypes of the study strains. (+), the gene is both present and in frame; (−), the gene is missing from a strain; (−)†, lgtG is present in 7946 but inactive because it has a mutation and is out of frame.
Mass Spectrometry of 7946 and 8024 LOS
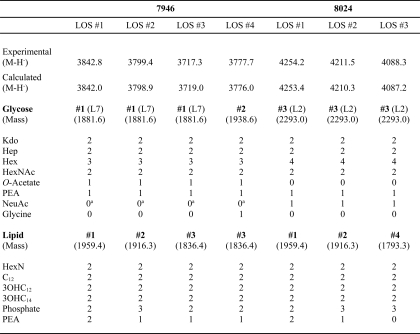
MALDI-TOF MS spectra of intact 7946 and 8024 LOS revealed that each strain made multiple lipid moieties to which a common glycose was linked (Table 4). The lipids differed in the number and positions of phosphate and PEA substitutions (phosphoforms). With one exception, masses of the glycose moieties were as expected from their immunotypes and genotypes; the compositions assigned to molecular ion peaks are shown in Table 4. The intact 7946 spectrum found two molecular ion peaks that had the same glycose compositions but differed in their lipid components. The highest mass peak, denominated LOS #1, was observed at m/z 3842.8 and was assigned the composition of Glycose #1 and Lipid #1. A second molecular ion peak, denominated LOS #2, was found at m/z 3799.4 and was assigned the composition of Glycose #1 and Lipid #2. Prompt fragment ions observed at m/z 1880.7 (Glycose #1), m/z 1938.4 (Glycose #2), m/z 1958.2 (Lipid #1), and m/z 1915.4 (Lipid #2) confirmed the assigned compositions.
TABLE 4.
Composition of molecular ion peaks in MALDI-TOF MS spectra of intact LOS
a Although not observed in MALDI-TOF MS spectra of intact LOS, (M-H−) peaks that resulted from the addition of N-acetylneuraminic (sialic) acid were seen in spectra of O-deacylated LOS.
An additional prompt fragment ion observed at m/z 1836.4 was assigned the composition of Lipid #3. A molecular ion peak consistent with Glycose #1 in combination with Lipid #3 was observed at m/z 3717.3 and denominated LOS #3. This ion peak was accompanied by an m/z 3777.7 molecular ion peak, denominated LOS #4, that would be consistent with the composition of Glycose #2 in combination with Lipid #3. Glycose #2 differs from Glycose #1 by the addition of glycine (m/z +57). This assignment was confirmed by the presence of a prompt fragmentation ion at m/z 1938.4. Molecular ion peaks that would be consistent with Glycose #2 in combination with either Lipid #1 or Lipid #2 were not seen.
The intact 8024 spectrum was more complex. Two glycoses that differed by the presence or absence of sialic acid (292.3 Da) were each attached to one of the four lipids in Table 4. These four sialylated LOS molecular ion peaks were at m/z 4254.2, m/z 4211.5, m/z 4131.5 (not shown in Table 4), and m/z 4088.3; the respective m/z −291.3 ion peaks were observed at m/z 3964.6, m/z 3919.6, m/z 3839.8, and m/z 3796.4. Prompt fragment ions consistent with the masses of the four respective lipid moieties were observed at m/z 1958.2, m/z 1915.4, m/z 1836.4, and m/z 1794.3, and a fragment ion consistent with Glycose #3 was seen at low abundance at m/z 2292.5. The mass of Glycose #3 was consistent with the structure of the L2 glycose in Table 2 but lacked the γ GlcNAc O-acetate.
As expected from its antigenic structure, a molecular ion peak consistent with the 7946 LOS #3 was seen at m/z 3717.3 along with a prompt fragment ion at m/z 1880.7 that would be consistent with the 7946 Glycose #1. Fragment ion peaks that represented various combinations of losses of Kdo (−220 Da), phosphate (−80 Da), and hexose (−162 Da) from the various molecules also were present.
Binding of LOS IgG to 7946 and 8024
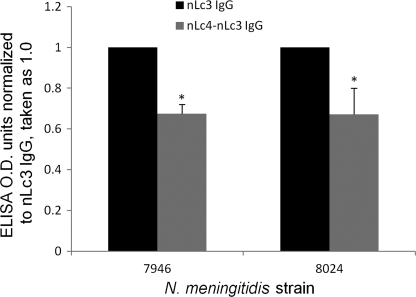
Both 7946 and 8024 cells bound nLc4-nLc3 IgG and nLc3 IgG (Fig. 3), but both strains bound significantly less nLc4-nLc3 IgG than nLc3 IgG, as would be expected, because the nLc3 IgG contained at least three specificities absent from the nLc4-nLc3 IgG. 8024 sialylates its LOS much more than does 7946, but this did not appear to affect the binding of nLc4-nLc3 IgG, because the decrease in binding of nLc4-nLc3 IgG, relative to that of nLc3 IgG, was the same for both strains.
FIGURE 3.
Binding of nLc3 (black bars) and nLc4-nLc3 (gray bars) LOS IgG to N. meningitidis strains, 7946 and 8024. Each IgG was tested at a concentration of 10 μg/ml. y axes are ELISA OD units normalized to the binding of nLc3 LOS IgG, which was assigned a value of 1.0. Error bars, S.D.; *, difference between the binding of nLc4-nLc3 IgG and nLc3 IgG to each strain was significant (p < 0.05; t test).
Bactericidal Activity of Human LOS IgG
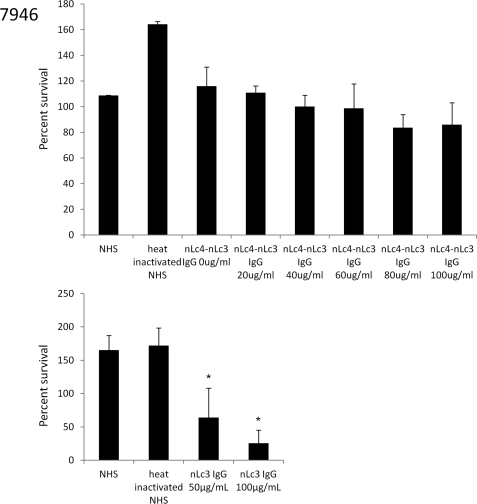
Both nLc4 IgG and nLc3 IgG killed 7946, but nLc4-nLc3 IgG did not kill at concentrations of 20–100 μg/ml (Fig. 4). Thus, all of the bactericidal activity in the nLc4 IgG was initiated by its nLc3 component. Increasing the concentration of nLc4-nLc3 IgG up to 100 μg/ml did not result in a significant reduction in survival of 7946 (Fig. 4).
FIGURE 4.
Bactericidal activity of nLc4-nLc3 IgG (top) and nLc3 IgG (bottom) for the L3,7 N. meningitidis strain, 7946. y axis, percentage of cfu present at time 0 that survived after 60 min in the presence of increasing concentrations of each IgG added to a human serum (NHS) used as the complement source. The NHS added 2.7 μg of nLc3 IgG and a negligible amount of nLc4-nLc3 IgG to each reaction. Error bars, S.D. values; *, p < 0.05 (t test; one tail) as compared with NHS. The nLc4-nLc3 IgG did not significantly reduce survival at any IgG concentration, whereas 50 μg/ml of the nLc3 IgG reduced survival by ∼50%, and 100 μg/ml of this IgG reduced survival by ∼75%.
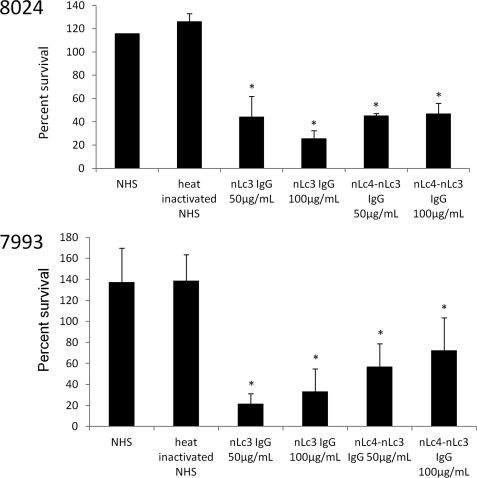
In contrast, nLc4-nLc3 IgG reduced survival of 8024 and 7993 by ∼60% at both 50 and 100 μg/ml IgG concentrations (Fig. 5). nLc3 IgG was somewhat more bactericidal, reducing survival by about 80% at a concentration of 100 μg/ml.
FIGURE 5.
Bactericidal activity of nLc4-nLc3 IgG and nLc3 IgG for N. meningitidis strains 8024 (L2,3) and 7993 (L2,4,6,7). y axis, percentage of cfu present at time 0 that survived after 60 min in the presence of 50 or 100 μg/ml of each IgG added to a human serum (NHS) used as the complement source. The NHS added 2.7 μg of nLc3 IgG to each of the 7993 reactions and 0.9 μg to each of the 8024 reactions but only a negligible amount of nLc4-nLc3 IgG to each reaction. Error bars, S.D. values; *, p < 0.05 (t test; one tail) as compared with NHS. The nLc4-nLc3 IgG at a concentration of 50 μg/ml significantly reduced survival of both strains, whereas it was unable to reduce survival of strain 7946 at even higher IgG concentrations. Increasing the IgG concentration to 100 μg/ml did not increase the killing of either strain. The nLc3 IgG also significantly reduced survival of both strains and was more bactericidal than the nLc4-nLc3 IgG at concentrations of 100 μg/ml.
7946 LOS Mutants
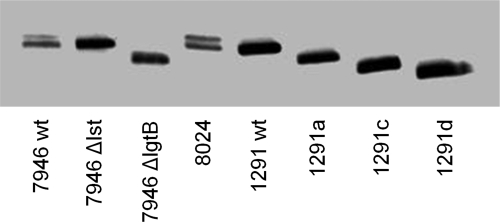
The 7946Δlst and 7946ΔlgtB mutants were designed to have the same LOS glycose as 1291wt and 1291a, respectively, but with the heterogeneous meningococcal lipids. PCR of the mutants with the primers in Table 3 confirmed the deletions, and as expected, migration of 7946Δlst and 7946ΔlgtB LOS in SDS-PAGE was equivalent to that of 1291 and 1291a, respectively (Fig. 6).
FIGURE 6.
SDS-PAGE separated LOS from 7946 and its Δlst and ΔlgtB mutants compared with LOS from 8026 and 1291wt, 1291a, 1291c, and 1291d. LOS were electrophoresed through 27 cm of 13.1% polyacrylamide gel and visualized with silver stain. 7946 and 8026 both make two LOS; the slower migrating molecule (upper band) is the sialylated LOS, and this glycoform is absent from 7946Δlst, which has the same electrophoretic mobility as 1291wt. 7946ΔlgtB has the same electrophoretic mobility as 1291a and migrates more slowly than 1291c.
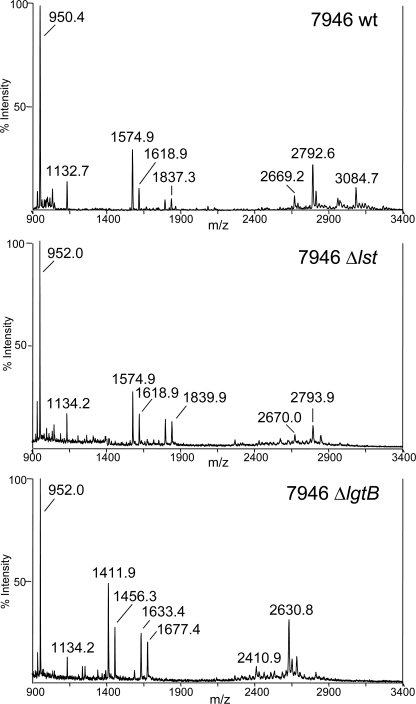
Negative ion MALDI-TOF MS of O-deacylated 7946wt, 7946Δlst and 7946ΔlgtB LOS confirmed the absence of the Lst- and LgtB-catalyzed glycoses, respectively, without additional structural alterations (Fig. 7). Prominent molecular ion peaks in the 7946wt LOS spectrum at m/z 3084.7 and m/z 2792.6 are in accordance with calculated masses (m/z 3083.8 and m/z 2792.6) for an O-deacylated LOS with a diphosphorylated lipid with one PEA and an nLc4 α chain, with and without sialic acid, respectively. These ions confirm the presence of the sialylated L3 and unsialylated L7 LOS glycoforms; the absence of the m/z 3084.7 ion in the spectrum of the 7946Δlst LOS confirms that lst is inactive in the mutant.
FIGURE 7.
MALDI-TOF mass spectra of O-deacylated LOS of 7946, 7946Δlst, and 7946ΔlgtB. Two prominent molecular ion peaks in the 7946wt LOS spectrum (top) at m/z 3084.7 and 2792.6 are in accordance with calculated masses (m/z 3083.8 and m/z 2792.6) for a diphosphorylated O-deacylated phosphoethanolaminylated LOS with an nLc4 α chain, with and without sialic acid substitution, respectively. The m/z 3083.8 peak is absent from the 7946Δlst spectrum. The m/z 2630.8 ion peak in the 7946ΔlgtB spectrum represents the loss of hexose (162 Da).
7946ΔlgtB LOS was expected to differ from 7946wt LOS by the absence of the nLc4 non-reducing terminal Gal (162 Da), as shown in Table 1. This was confirmed by the mass difference of 163.1 Da between the molecular ions at m/z 2630.8 in the 7946ΔlgtB spectrum and the molecular ions at m/z 2793.9 in the 7946Δlst spectrum. The loss of hexose was supported further by the oligosaccharide fragment ions in the 7946ΔlgtB LOS spectrum at m/z 1677.4 and m/z 1456.3 that differ from the analogous ions in the 7946Δlst LOS spectrum at m/z 1839.9 and m/z 1618.9, respectively, by 162 Da.
Use of the reflectron mass analyzer enabled assignment of oligosaccharide fragment ion masses with greater mass accuracy. For example, using external calibration, the m/z 1618.9 ion in Fig. 7 was calculated at m/z 1618.410 in the 7946Δlst LOS spectrum; this differs from the calculated mass of 1618.503 by ∼60 ppm. The 1618.5 Da ion represents the L7 glycose minus one Kdo (220 Da) and the γ chain GlcNAc O-acetate (42 Da). Higher mass ions were not analyzed.
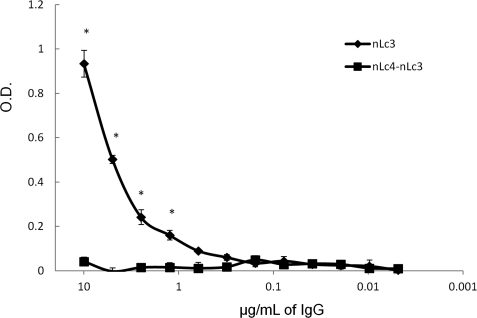
Binding of LOS IgG to 7946ΔlgtB
Because the 7946ΔlgtB mutant lacks the nLc4 non-reducing terminal Gal residue, it should not bind nLc4-nLc3 IgG, and it did not (Fig. 8). As expected, nLc3 IgG, which should not require this nLc4 Gal residue for binding, bound the 7946ΔlgtB mutant (Fig. 8). This confirmed the specificity of the nLc4-nLc3 IgG for the nLc4 terminal Gal.
FIGURE 8.
Binding of nLc4-nLc3 (■—■) and nLc3 (♦—♦) IgG to 7946ΔlgtB. Error bars, S.D. values; *, p < 0.05 (t test; one tail) for the binding of nLc3 IgG as compared with nLc4-nLc3 IgG.
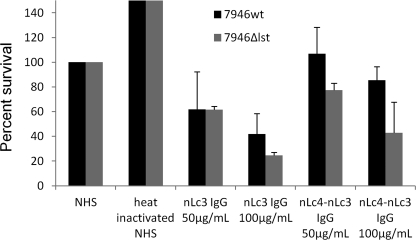
Bactericidal Activity of LOS IgG for 7946Δlst and 7946ΔlgtB
nLc3 IgG, at concentrations of 50 and 100 μg/ml, reduced survival of 7946Δlst by 39 and 76%, respectively (Fig. 9). The corresponding values for nLc4-nLc3 IgG were 23 and 58%. The ΔlgtB 7946 mutant did not survive in the serum used as a complement source.
FIGURE 9.
Bactericidal activity for 7946wt and its Δlst mutant of human nLc4-nLc3 and nLc3 IgG. y axis, percentage of cfu present at time 0 that survived after 60 min in the presence of 50 or 100 μg/ml of each IgG added to a human serum (NHS) used as the complement source. Data are normalized to survival in NHS, assigned a value of 100%. Error bars, S.D. values. The NHS added 0.9 μg of nLc3 IgG and a negligible amount of nLc4-nLc3 IgG to each reaction. nLc4-nLc3 IgG reduced survival of 7946Δlst (p = 0.11 for the difference in survival between 7946wt and 7946Δlst in 50 μg/ml nLc4-nLc3 IgG, and p = 0.07 for the difference in 100 μg/ml nLc4- nLc3 IgG).
DISCUSSION
These experiments provide new insights into the immunochemistry of neisserial LOS. Each LOS glycoform was thought to present a single antigen centered on the non-reducing terminal glycose, the “immunodominant glycose,” as modified by the various substitutions of Hep2. This paradigm informs the LOS serotyping system (16–20) and many previous studies, including some of our own (12–14). The diversity of LOS antibodies, according to this notion, was provided by the diversity of LOS glycoforms. The finding that the nLC4 LOS α chain conforms four or more different antigens was not expected and leads us to conclude that the diversity of LOS antibodies is as much a function of the presentation of multiple antigens within the α chain as it is of the presence of multiple different glycoforms.
Both 1291d and 1291e have mutations in pgm, which encodes the neisserial phosphoglucomutase, and neither can convert glucose 6-phosphate to glucose 1-phosphate (27). Because the LOS α chain is initiated by β-Glc, pgm mutations would be expected to result in the absence of an α chain. However, both the 1291d and 1291e pgm mutations occur outside the active site involved in phosphate binding, both mutants are thought to be able to take up and use glucose 1-phosphate from the growth medium (27), and both have been shown to be able to make some LOS molecules with an α chain β-Glc (15). As can be seen in Fig. 1, the 1291e mutant used in this study made two LOS species that migrated closely together in a slower and broader band than the 1291d LOS. We conclude that this 1291e mutant, but not the 1291d mutant, did make a small amount of LOS with an α chain β-Glc and that this structure represents a fourth α chain antigen. In the original description of the mutants, 1291d migrated more slowly than 1291e and therefore received the “d” designation (23).
A Δpgm mutant that does not acquire exogenous glucose 1-phosphate, such as 1291d, has the same phenotype as a ΔlgtF mutant; their LOS consists of a complex basal glycose and the lipid moiety. The 1291d basal glycose is composed of a phosphoethanolaminyl diheptoside, two dOclA residues, and the γ chain GlcNAc. This complex glycolipid probably forms multiple antigens that are not conformed by the α chain, so the number of antigens conformed by 1291wt LOS is almost certainly greater than our estimate. Roughly half of the nLc4 IgG recognized one or more basal antigens, as shown by binding to 1291d. We did not seek to understand the antigens presented by Δpgm/ΔlgtF mutants.
Partially hydrolyzed LOS have a single lipid phosphoform (44), so the finding of multiple phosphoforms also was unexpected. MS of intact LOS of the group C strain, 89I, did find multiple lipid phosphoforms (42, 43), but this might have been an exception. What effects, if any, the different lipids might have on glycose antigen formation (48, 49) were not explored.
Also surprising was that the nLc4 terminal Gal appeared to protect L3,7 but not L2,4 strains from bactericidal activity. The ΔlgtB mutant, which lacks this terminal Gal, was unable to survive in the serum used as a complement source. The nLc4-nLc3IgG that required the terminal Gal for binding was not able to overcome its protective effect for L3,7 strains. This IgG did not bind the 7946ΔlgtB mutant that has the nLc3 α chain and the same lipids as the parent strain. Thus, we can conclude that it was specific for the nLc4 terminal Gal and that its specificity was not influenced by basal adornments or its lipid. The sensitivity of L2,4 strains to bacteriolysis by nLc4-nLc3 IgG could explain why they cause endemic disease less frequently (10–13).
LOS synthesis by meningococci follows that by gonococci (45), except meningococci do not have an active LgtD. L3 and L7 strains substitute C3 of Hep2 with PEA (16, 18), catalyzed by Lpt3 (34), whereas L2, L4, and L6 strains substitute C6 or C7 of Hep2 with PEA (16, 17, 20), catalyzed by Lpt6 (35). L2 strains substitute C3 of Hep2 with α-glucose (α-Glc) (17), catalyzed by LgtG (33). An additional β-GlcNac basal substitution has been found on L1 strains (12), and L3 and L4 strains have been reported to have glycine at C7 of Hep2 (21). We did not explore how these various substitutions affect sensitivity to the bactericidal activity of the two IgG preparations.
40% of 8024 and 7993 bacteria survived 100 μg/ml concentrations of nLc4-nLc3 IgG. Both strains make the L3,7 as well as the L2 glycoforms. The genetic basis of the synthesis of the two glycoforms is such that they are mutually exclusive, and the bacteria making each represents a distinct subpopulation of the global strain population, as has been shown to be the case for both N. gonorrhoeae (24, 50) and N. meningitidis (51) glycotypes within a strain population. We conclude that the surviving bacteria were the intrinsically resistant L3,7 subpopulations of each strain.
Resistance of L3,7 bacteria to killing by nLc4-nLc3 IgG could not be explained by poor binding because it bound to these strains almost as well as did the nLc3 IgG. Nor could it be explained by any intrinsic defect in its ability to activate complement, because it was bactericidal for L2,4 strains. Surprisingly, LOS sialylation did not affect the binding of either IgG, because binding to the heavily sialylated 8024 bacteria was the same as that to the meagerly sialylated 7946 bacteria.
Bacteria may survive in serum because they intrinsically resist killing by the antibodies present or because the concentration of potentially bactericidal antibodies is inadequate. The ratio of IgG to bacteria in the assay is skewed in favor of survival; 100–300 cfu of bacteria are exposed to μg of IgG in a fixed volume of serum that is not renewed by circulation. IgG specific for the group C meningococcal capsule is bactericidal at concentrations of <2 μg/ml, whereas survival of 7946 was not significantly reduced by exposure to 100 μg/ml of nLc4-nLc3 IgG. We conclude that the resistance of 7946 to killing initiated by the nLc4-nLc3 IgG was intrinsic and not due to inadequate concentrations of IgG. At high concentrations of IgG, some cross-linking of the bacteria occurs, which can spuriously diminish the observed cfu. In contrast with nLc4-nLc3 IgG, IgG specific for one or more of the internal nLc3 antigens efficiently killed both L3,7 and L2,4 strains, and intrinsic resistance to killing by nLc3 IgG was not observed.
Why IgG binding to the nLc4 α chain of the L3,7 and L2,4 glycoforms should result in such different outcomes is not clear. IgG propagates complement activation through the classical pathway C3 convertase, C4b2a. The Hep2 PEA substitutions provide primary NH2 groups with which C4b can form amide linkages. The exocyclic PEA of the L2,4 glycoform binds C4b more efficiently than the cyclic PEA of the L3,7 glycoform (52) and thus should maximize the bactericidal effectiveness of bound IgG, but this does not explain the equal sensitivity of L2,4 and L3,7 strains to the bactericidal activity of the nLc3 IgG.
The exquisite sensitivity to usually fatal meningococcal disease of men with X-linked properdin deficiency and therefore absent alternative complement pathway (ACP) activity (53) shows the importance of the ACP in protection against meningococcal dissemination. When the ratio of IgG to inoculum is low, the density of C3b deposited on the bacterial surface through IgG activation of the classical pathway is not adequate to sustain bacterial lysis without ACP augmentation (54). LOS sialylation interferes with sustained ACP activation (47), and the 7946Δlst mutant, unlike its parent, was modestly sensitive to killing initiated by nLc4-nLc3 IgG. But the two L2 strains have more heavily sialylated nLc4 α chains than 7946, and they were killed by nLc4-nLc3 IgG. Furthermore, loss of LOS sialylation had little effect on killing by nLc3 IgG, so LOS sialylation also does not fully explain the difference in sensitivity to killing of L3,7 and L2,4 bacteria by nLc4-nLc3 IgG.
In summary, IgG specific for one or more of the internal nLc3 antigens was bactericidal for both L2,4 and L3,7 strains; their bactericidal effect was dependent on the concentration of IgG and was not affected by LOS sialylation. In contrast, IgG specific for the nLc4 terminal Gal was unable to kill bacteria that made the L3,7 glycoform but was bactericidal for those that made the L2,4 glycoform. Resistance of L3,7 strains to this IgG was mediated, in part, by LOS sialylation, whereas sialylation of L2,4 strains did not prevent their killing by the antibody. These data support the conclusion that induction of nLc3 IgG by N. lactamica colonization or vaccination during infancy would protect children from disseminated meningococcal disease.
Acknowledgment
Mingfeng Liu provided many helpful suggestions during preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 053728 and AI 065605, administered by the Northern California Institute for Research and Education (to J. M. G.) and AI 063927 (to G. A. J.). This work was also supported by a Veterans Affairs Merit Review Award (to G. A. J.) and with resources of the Veterans Affairs Medical Center (San Francisco, CA). This is publication 109 from the Center for Immunochemistry.
Designations for LOS glycoses follow the recommendations of the International Union of Pure and Applied Chemistry/International Union of Biochemistry and Molecular Biology (IUPAC-IUB) Joint Commission on Biochemical Nomenclature, Nomenclature of Glycolipids (1997). These recommendations can be found at the IUPAC-IUB Web site.
- LOS
- lipooligosaccharide(s)
- nLc4
- lacto-N-neotetraose
- nLc3
- lacto-N-neotriaose
- Lc2
- lactose
- Gb3
- globotriaose
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- PEA
- phosphoethanolamine
- Hep
- heptose
- NeuAc
- N-acetylneuraminic (sialic) acid
- SPRIA
- solid phase radioimmunoassay
- ANOVA
- analysis of variance
- NHS
- normal human serum
- ACP
- alternative complement pathway
- Kdo
- 3-deoxy-d-manno-octulosonic acid.
REFERENCES
- 1. Rosenstein N. E., Perkins B. A., Stephens D. S., Popovic T., Hughes J. M. (2001) New Engl. J. Med. 344, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 2. Goldschneider I., Gotschlich E. C., Artenstein M. S. (1969) J. Exp. Med. 129, 1307–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Griffiss J. M., Bertram M. A. (1977) J. Infect. Dis. 136, 733–739 [DOI] [PubMed] [Google Scholar]
- 4. Nicholson A., Lepow I. H. (1979) Science 205, 298–299 [DOI] [PubMed] [Google Scholar]
- 5. Gold R., Goldschneider I., Lepow M. L., Draper T. F., Randolph M. (1978) J. Infect. Dis. 137, 112–121 [DOI] [PubMed] [Google Scholar]
- 6. Goldschneider I., Gotschlich E. C., Artenstein M. S. (1969) J. Exp. Med. 129, 1327–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weber M. V., Claus H., Maiden M. C., Frosch M., Vogel U. (2006) Int. J. Med. Microbiol. 296, 475–484 [DOI] [PubMed] [Google Scholar]
- 8. Kasper D. L., Winkelhake J. L., Brandt B. L., Artenstein M. S. (1973) J. Infect. Dis. 127, 378–387 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. J., Mandrell R. E., Griffiss J. M. (1989) Infect. Immun. 57, 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Estabrook M. M., Baker C. J., Griffiss J. M. (1993) J. Infect. Dis. 167, 966–970 [DOI] [PubMed] [Google Scholar]
- 11. Estabrook M. M., Mandrell R. E., Apicella M. A., Griffiss J. M. (1990) Infect. Immun. 58, 2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McLeod Griffiss J., Brandt B. L., Saunders N. B., Zollinger W. (2000) J. Biol. Chem. 275, 9716–9724 [DOI] [PubMed] [Google Scholar]
- 13. Griffiss J. M., Brandt B. L., Broud D. D., Goroff D. K., Baker C. J. (1984) J. Infect. Dis. 150, 71–79 [DOI] [PubMed] [Google Scholar]
- 14. Estabrook M. M., Jarvis G. A., McLeod Griffiss J. (2007) Infect. Immun. 75, 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. John C. M., Griffiss J. M., Apicella M. A., Mandrell R. E., Gibson B. W. (1991) J. Biol. Chem. 266, 19303–19311 [PubMed] [Google Scholar]
- 16. Kogan G., Uhrín D., Brisson J. R., Jennings H. J. (1997) Carbohydr. Res. 298, 191–199 [DOI] [PubMed] [Google Scholar]
- 17. Gamian A., Beurret M., Michon F., Brisson J. R., Jennings H. J. (1992) J. Biol. Chem. 267, 922–925 [PubMed] [Google Scholar]
- 18. Pavliak V., Brisson J. R., Michon F., Uhrín D., Jennings H. J. (1993) J. Biol. Chem. 268, 14146–14152 [PubMed] [Google Scholar]
- 19. Zollinger W. D., Mandrell R. E. (1977) Infect. Immun. 18, 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Fabio J. L., Michon F., Brisson J. R., Jennings H. J. (1990) Can. J. Chem. 68, 1029–1034 [Google Scholar]
- 21. Cox A. D., Li J., Richards J. C. (2002) Eur. J. Biochem. 269, 4169–4175 [DOI] [PubMed] [Google Scholar]
- 22. Rahman M. M., Stephens D. S., Kahler C. M., Glushka J., Carlson R. W. (1998) Carbohydr. Res. 307, 311–324 [DOI] [PubMed] [Google Scholar]
- 23. Apicella M. A. (1976) J. Infect. Dis. 134, 377–383 [DOI] [PubMed] [Google Scholar]
- 24. Schneider H., Griffiss J. M., Boslego J. W., Hitchcock P. J., Zahos K. M., Apicella M. A. (1991) J. Exp. Med. 174, 1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamasaki R., Bacon B. E., Nasholds W., Schneider H., Griffiss J. M. (1991) Biochemistry 30, 10566–10575 [DOI] [PubMed] [Google Scholar]
- 26. Dudas K. C., Apicella M. A. (1988) Infect. Immun. 56, 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandlin R. C., Danaher R. J., Stein D. C. (1994) J. Bacteriol. 176, 6869–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gotschlich E. C. (1994) J. Exp. Med. 180, 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Danaher R. J., Levin J. C., Arking D., Burch C. L., Sandlin R., Stein D. C. (1995) J. Bacteriol. 177, 7275–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider H., Hale T. L., Zollinger W. D., Seid R. C., Jr., Hammack C. A., Griffiss J. M. (1984) Infect. Immun. 45, 544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider H., Griffiss J. M., Williams G. D., Pier G. B. (1982) J. Gen. Microbiol. 128, 13–22 [DOI] [PubMed] [Google Scholar]
- 32. Yang Q. L., Gotschlich E. C. (1996) J. Exp. Med. 183, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banerjee A., Wang R., Uljon S. N., Rice P. A., Gotschlich E. C., Stein D. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10872–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mackinnon F. G., Cox A. D., Plested J. S., Tang C. M., Makepeace K., Coull P. A., Wright J. C., Chalmers R., Hood D. W., Richards J. C., Moxon E. R. (2002) Mol. Microbiol. 43, 931–943 [DOI] [PubMed] [Google Scholar]
- 35. Wright J. C., Hood D. W., Randle G. A., Makepeace K., Cox A. D., Li J., Chalmers R., Richards J. C., Moxon E. R. (2004) J. Bacteriol. 186, 6970–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandrell R. E., Kim J. J., John C. M., Gibson B. W., Sugai J. V., Apicella M. A., Griffiss J. M., Yamasaki R. (1991) J. Bacteriol. 173, 2823–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamasaki R., Griffiss J. M., Quinn K. P., Mandrell R. E. (1993) J. Bacteriol. 175, 4565–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilbert M., Watson D. C., Cunningham A. M., Jennings M. P., Young N. M., Wakarchuk W. W. (1996) J. Biol. Chem. 271, 28271–28276 [DOI] [PubMed] [Google Scholar]
- 39. Hitchcock P. J., Brown T. M. (1983) J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swanson K. V., Griffiss J. McL. (2006) Carbohydr. Res. 341, 388–396 [DOI] [PubMed] [Google Scholar]
- 41. Apicella M. A., Griffiss J. M., Schneider H. (1994) Methods Enzymol. 235, 242–252 [DOI] [PubMed] [Google Scholar]
- 42. John C. M., Liu M., Jarvis G. A. (2009) J. Lipid Res. 3, 424–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. John C. M., Liu M., Jarvis G. A. (2009) J. Biol. Chem. 284, 21515–21525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kulshin V. A., Zähringer U., Lindner B., Frasch C. E., Tsai C. M., Dmitriev B. A., Rietschel E. T. (1992) J. Bacteriol. 174, 1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kahler C. M., Stephens D. S. (1998) Crit. Rev. Microbiol. 24, 281–334 [DOI] [PubMed] [Google Scholar]
- 46. Bertram M. A., Griffiss J. M., Broud D. D. (1976) J. Immunol. 116, 842–846 [PubMed] [Google Scholar]
- 47. Estabrook M. M., Griffiss J. M., Jarvis G. A. (1997) Infect. Immun. 65, 4436–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamasaki R., Schneider H., Griffiss J. M., Mandrell R. E. (1988) Mol. Immunol. 25, 799–809 [DOI] [PubMed] [Google Scholar]
- 49. Kim J. J., Phillips N. J., Gibson B. W., Griffiss J. M., Yamasaki R. (1994) Infect. Immun. 62, 1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schneider H., Hammack C. A., Apicella M. A., Griffiss J. M. (1988) Infect. Immun. 56, 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Estabrook M. M., Hyun W. C., Griffiss J. M. (1992) Abstracts of the 92nd Annual Meeting of the American Society for Microbiology, New Orleans, LA, 26–30 May, p. 43, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 52. Ram S., Cox A. D., Wright J. C., Vogel U., Getzlaff S., Boden R., Li J., Plested J. S., Meri S., Gulati S., Stein D. C., Richards J. C., Moxon E. R., Rice P. A. (2003) J. Biol. Chem. 278, 50853–50862 [DOI] [PubMed] [Google Scholar]
- 53. Densen P., Weiler J. M., Griffiss J. M., Hoffmann L. G. (1987) New Engl. J. Med. 316, 922–926 [DOI] [PubMed] [Google Scholar]
- 54. Griffiss J. M., Jarvis G. A., O'Brien J. P., Eads M. M., Schneider H. (1991) J. Immunol. 147, 298–305 [PubMed] [Google Scholar]