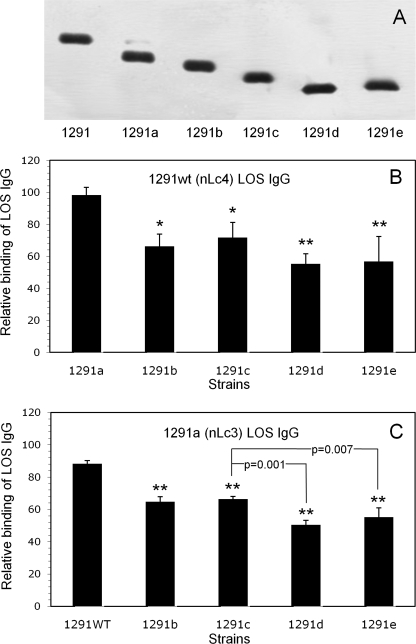
FIGURE 1.
Binding of affinity-purified 1291wt (nLc4) LOS IgG and 1291a (nLc3) LOS IgG to 1291 and its pyocin-selected mutants. A, SDS-PAGE separated LOS from 1291 and each of the 1291 mutants. LOS were electrophoresed through 27 cm of 13.1% polyacrylamide gel and visualized with silver stain. The LOS α chain structures are in Table 1. Note that 1291a makes a small amount of 1291wt LOS and that 1291e LOS is composed of two closely migrating species that, together, migrate more slowly than 1291d. B, whole-cell ELISA of 1291wt (nLc4) LOS IgG. Microtiter wells were coated with the strains and then reacted with 10 μg/ml IgG. Binding is expressed as the percentage of the binding of the IgG to 1291wt (100%). Error bars, S.D. of three assays. Differences in binding to the strains were statistically significant (p = 0.002; ANOVA). Pairwise multiple comparisons (Student-Newman-Keuls method) revealed that the 1291b–e mutants bound significantly less nLc4 LOS IgG than 1291a (**, p = 0.002; *, p = 0.007). C, whole-cell ELISA of 1291a (nLc3) LOS IgG. Microtiter wells were coated with the strains and then reacted with 10 μg/ml IgG. Binding is expressed as a percentage of the binding of the IgG to 1291a (100%). Error bars, S.D. of three assays. Differences in binding to the strains were statistically significant (p < 0.001; ANOVA). Pairwise multiple comparisons revealed not only that the 1291b–e mutants bound significantly less nLc3 LOS IgG than 1291wt (**, p < 0.001) but that both 1291d (p = 0.001) and 1291e (p = 0.007) bound less than 1291c or 1291b. The difference in binding between 1291d and 1291e was not significant (p = 0.12).