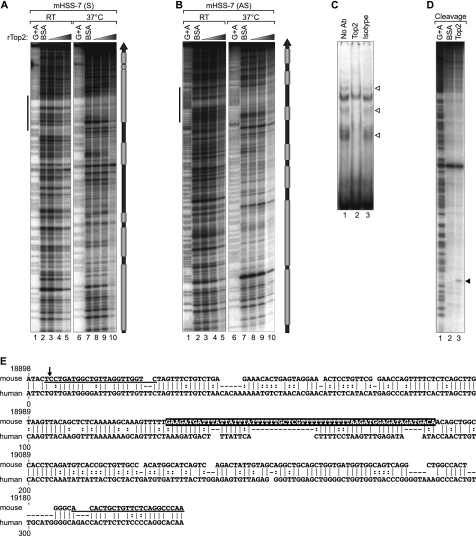
FIGURE 6.
HSS−7 is a substrate for Top2 in vitro. A and B, quantitative DNase I footprinting analysis of Top2 binding to HSS−7. 308-bp double-stranded DNA probes containing murine HSS−7 radiolabeled on the sense (mHSS−7 (S); A) and antisense (mHSS−7 (AS); B) strands were incubated with increasing concentrations of recombinant human Top2 protein (rTop2) at room temperature (RT) or at 37 °C. DNase I-digested probes were resolved by 8% denaturing PAGE and subjected to autoradiography. Gray boxes to the right of A and B denote Top2-binding positions. C, EMSA performed by incubating a 64-bp HSS−7 probe (sequence highlighted in E, with its position indicated by vertical bars to the left of A and B) with nuclear extracts from LPS-stimulated J774 cells. The EMSA was performed with an isotype control or anti-Top2 antibody; disrupted Top2 complexes are indicated by arrowheads. D, concentrated and purified recombinant Drosophila Top2 (lane 3) was utilized in an in vitro cleavage reaction with 308-bp radiolabeled DNA containing the murine HSS−7 region and resolved by 8% denaturing PAGE. E, PipMaker alignment of mouse (308 bp) and human (288 bp) HSS−7 sequences. Dashes represent gaps in the alignment, vertical lines represent aligned identical nucleotides, and dots indicate a purine or pyrimidine change in the sequence. The numbers at the top of the alignment correspond to positions of the mouse sequence in the TNF locus as in Fig. 2. The sequences of primers used in MAR and ChIP assays are underlined.