Figure 6.
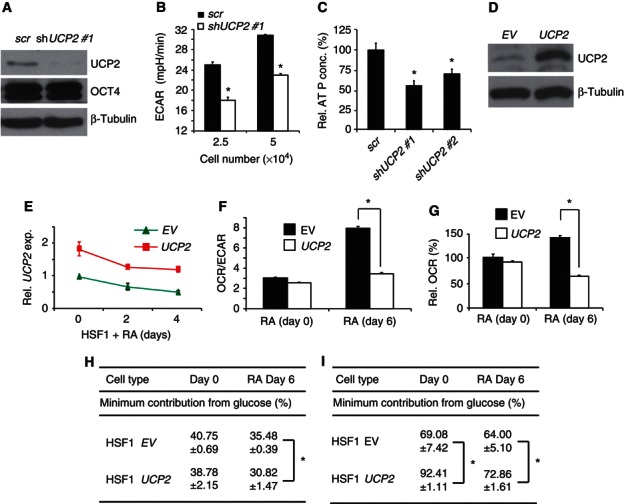
UCP2 regulates energy metabolism in hPSCs and during differentiation. (A) Western blots in HSF1 cells transduced with shUCP2 #1 and a scramble (scr) control. (B) ECAR for shUCP2 #1 and scr HSF1 cells (n=3), * P<0.05. (C) ATP normalized for HSF1 cell number with shUCP2 knockdown (n=3), * P<0.05. (D) UCP2 immunoblot in HSF1 cells transduced with empty vector and UCP2‐expressing lentiviruses. (E) UCP2 expression by QPCR in EV and UCP2 transduced HSF1 cells with 10 μM RA‐induced differentiation for up to 4 days (n=3). (F, G) OCR/ECAR and OCR for EV or UCP2 transduced HSF1 cells exposed to 10 μM RA for up to 6 days (n=3–7), * P<0.005. (H) Stable isotope tracing analysis. hPSCs were grown in StemPro SFM medium (17.5 mM glucose) supplemented with 7.5 mM [U13C6]‐labelled glucose for 48 h. ∑mn (average 13C/molecule) was calculated from the mass isotopomer distribution in glutamate. Minimum contribution from glucose was calculated through dividing the observed ∑mn glutamate by the theoretical ∑mn glutamate (7.5/25 × 4), when all glucose carbons become ∑mn carbons in the C2–C5 fragment of glutamate. Data are expressed as mean±s.d. (n=3), * P<0.02. (I) Stable isotope tracing analysis with cells grown as in (H), ∑mn was calculated from the mass isotopomer distribution in ribose. The minimum contribution from glucose was calculated through dividing the observed ∑mn ribose by the theoretical ∑mn ribose (7.5/25 × 5). Data are expressed as mean±s.d. (n=3), * P<0.05.
