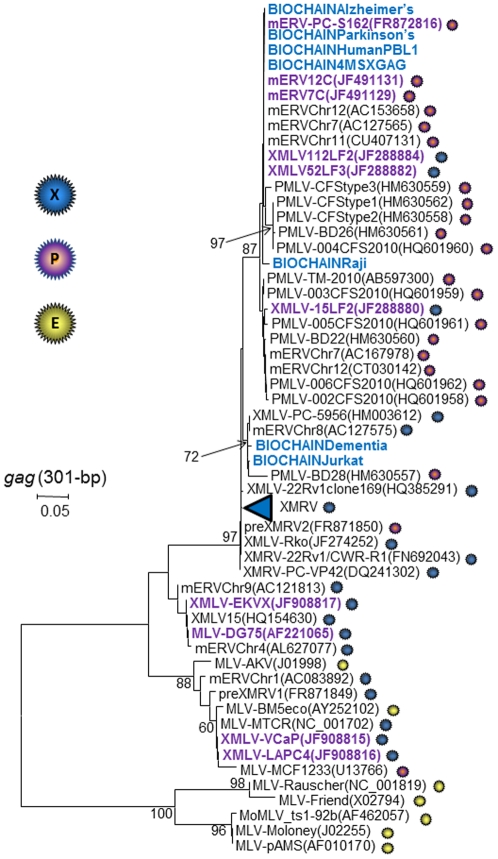
Figure 4. Inference of contamination origin in commercial human genomic DNAs.
Phylogenetic analysis of 301-bp gag sequences. Bootstrap values ≥60 are shown. New sequences from the current study are shown in blue (human genomic DNA contaminants). Sequence names in purple are those reported as contaminants from other human studies. GenBank accession numbers for prototypical murine leukemia virus (MLV) reference sequences are provided in parentheses. XMRV clades are collapsed for presentation only and include CFS-WPI-1106(GQ497344), CFS-WPI-1178(GQ497343), CFS-WPI-1130(GQ483508), CFS-WPI-1138(GQ483509), CFS-WPI-1169(GQ483510), CFS-WPI-CI-1303(JF907633), CFS-WPI-CI-1313(JF907643), CFS-WPI-CI-1314T(JF907644), CFS-WPI-CI-1307(JF907638), CFS-WPI-CI-1310(JF907641), CFS-WPI-CI-1327(JF907636), preXMRV2(FR871850), PC-VP35(DQ241301),and PC-VP62(DQ399707). Accession numbers for the new gag sequences generated in our study are JN629081-JN629087. Sequences coded as XMRV VP and WPI are from prostate cancer (PC) and CFS patients, respectively. BD; blood donor sequences. Viral host receptor tropism is indicated by blue (xenotropic), purple (polytropic), and yellow (ecotropic) spheres. PMLV, polytropic MLV; XMLV, xenotropic MLV.