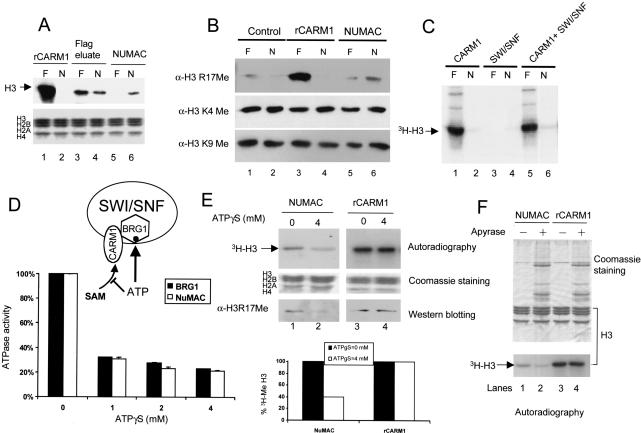
Figure 2.
NUMAC ATPase activity is required for nucleosomal histone methylation. (A) Nucleosomal histone methylation activity of 0.2 μg of rCARM1 (lanes 1,2), 0.2 μg of Flag-peptide eluates (lanes 3,4), or 0.1 μg of NUMAC (Fraction 40, lanes 5,6) were determined with 3 μg of either free core histones (F) or nucleosomes (N) in the methylation reaction. Autoradiograph (top) shows 3H-H3 in nucleosomes after methylation, and Coomassie staining (bottom) shows total core histones. (B) Determination of methylation sites in free core histones (F) and nucleosomes (N) by rCARM1 (lanes 3,4) and NUMAC (lanes 5,6). A total of 3 μg of free core histones (lanes 1,3,5) or nucleosomes (lanes 2,4,6) were incubated with 0.1 μg of BSA (lanes 1,2), 0.1 μg of rCARM1 (lanes 3,4), or 0.1 μg of NUMAC (lanes 5,6) in the presence of 1 mM s-AdoMet. Reaction products were resolved by SDS-PAGE and analyzed using anti-H3R17Me antibodies. The blot was stripped and reprobed with anti-H3K4Me or H3K9Me antibodies to indicate equal loading and the unaltered methylation at those sites. (C) Addition of rCARM1 to SWI/SNF minimally affects nucleosomal methylation. A total of 0.1 μg of rCARM1 (lanes 1,2), 3 μg of SWI/SNF (lanes 3,4), or 0.1 μg of rCARM1 preincubated with 3 μg of SWI/SNF (lanes 5,6) at 27°C for 15 min were incubated with 3 μg of either free core histones (lanes 1,3,5) or nucleosomes (lanes 2,4,6) in the methylation reaction. 3H-H3 is shown in the autoradiograph. (D) Dual role of ATP on NUMAC and determination of ATPγS concentration required to inhibit BRG1-ATPase activity. A total of 0.05 μg of BRG1 or 0.5 μg of NUMAC were incubated with [γ-32P]ATP and 0-4 mM ATP in the ATPase assay. Free phosphate was separated from [γ-32P]ATP on TLC plates, and the percentage of hydrolyzed [γ-32P]ATP was plotted. (E) Inhibition of NUMAC nucleosome (lanes 1,2) but not rCARM1 histone (lanes 3,4) methylation by 4 mM ATPγS. A total of 0.5 μg of NUMAC or 0.1 μg of rCARM1 was incubated with 3 μg of nucleosomes or free core histones, respectively, in the presence of 0.1 mM 3H-AdoMet. Autoradiography and Coomassie staining of SDS-PAGE are shown. Half of the reactions were subjected to Western blotting using α-H3R17Me antibody. The 3H-histone H3 band was cut out and counted with scintillation solvent. The amount of 3H-histone H3 in the presence of 4 mM ATPγS (lanes 2,4) was normalized by the level without ATPγS (lanes 1,3) to show the inhibitory effect of ATPγS (bottom). (F) Either 0.5 μg of NUMAC or 0.1 μg of rCARM1 were used to methylate 3 μg of nucleosome or free core histones, respectively. A 0.2 unit of apyrase was preincubated with the reaction mixture at 27°C for 15 min to deplete ATP before addition of 3H-AdoMet substrates.