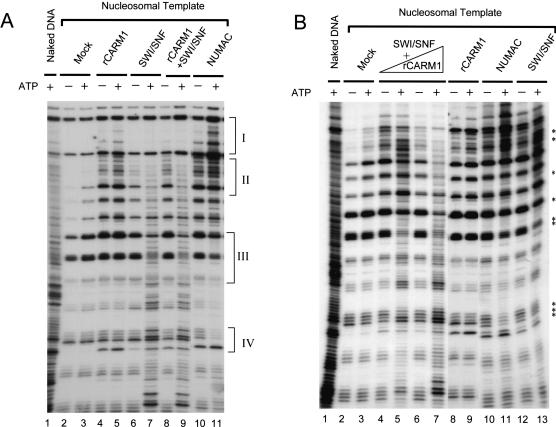
Figure 3.
Chromatin-remodeling activity of NUMAC and stimulation of SWI/SNF-remodeling activity by CARM1. (A) NUMAC and SWI/SNF exhibit different nucleosome disruption patterns. The ATP-dependent remodeling as revealed by DNaseI digestion of mononucleosomes after incubating with mock (0.1 μg of BSA, lanes 2,3), rCARM1 (0.1 μg, lanes 4,5), SWI/SNF (0.6 μg, lanes 6,7), rCARM1+SWI/SNF (0.1 μg rCARM1 pre-incubated with 0.6 μg of SWI/SNF at 27°C for 15 min, which gave a molar ratio of 5:1 for rCARM1/SWI/SNF, lanes 8,9), NUMAC (0.6 μg, lanes 10,11). Brackets I and II highlight the ATP-dependent changes in DNaseI digestion pattern increased by NUMAC and decreased by SWI/SNF; brackets III and IV highlight the opposite effect generated by SWI/SNF or NUMAC. (B) Stimulation of SWI/SNF remodeling activity by rCARM1 at limiting concentrations of SWI/SNF. The ATP-dependent remodeling of mononucleosomes by mock (0.1 μg of BSA, lanes 2,3), rCARM1 (0.1 μg, lanes 8,9), SWI/SNF (0.12 μg, lanes 12,13), NUMAC (0.12 μg, lanes 10,11), and rCARM1 and SWI/SNF mix [6 ng of rCARM1 pre-incubated with 0.12 μg of SWI/SNF, lanes 4,5, or 30 ng of rCARM1 preincubated with 0.12 μg of SWI/SNF, lanes 6,7, which gave 1:1 (lanes 4,5) or 5:1 (lanes 6,7) molar ratio for rCARM1/SWI/SNF]. Stars denote weak ATP-dependent changes by SWI/SNF and NUMAC.