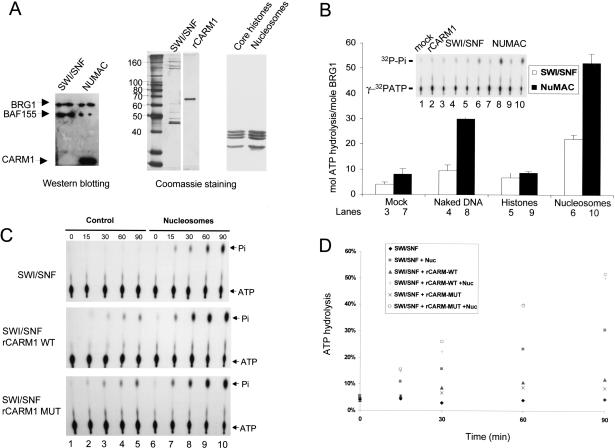
Figure 4.
CARM1 stimulates ATPase activity of NUMAC and BRG1. (A) Western normalization of BRG1 amount in SWI/SNF and NUMAC complex. Approximately 100 ng of SWI/SNF and NUMAC were loaded on 8% SDS-PAGE, transferred to nitrocellulose membrane, and blotted with the mixture of BRG1/BAF155/CARM1 antibodies (left). A total of 0.5 μg of SWI/SNF, 0.1 μg of rCARM1, 2 μg of core histones, and 3 μg of nucleosomes were resolved on SDS-PAGE and Coomassie stained (right). (B) Stimulation of SWI/SNF (0.3 μg, white) and NUMAC (0.3 μg, black) ATP hydrolysis by 0.1 μg of BSA (lanes 3,7), 1 μg of 1-kb plasmid DNA (lanes 4,8), 3 μg of core histones (lanes 5,9), or 3 μg of nucleosomes (lanes 6,10). The hydrolysis of 1.7 pmole [γ-32P]ATP by SWI/SNF and NUMAC in the presence of BSA (mock), naked DNA, histones, or nucleosomes was analyzed by TLC, and the autoradiography (inset) is shown. The hydrolysis of [γ-32P]ATP in the presence of 0.1 μg of BSA (lane 1) or 0.1 μg of rCARM1 (lane 2) were shown as controls. SWI/SNF and NUMAC are normalized to equimolar amounts of BRG1. The molar amount of ATP hydrolyzed by per mole of BRG1 in each complex is presented on the y-axis. (C) CARM1 stimulates ATPase activity of SWI/SNF. Time course of [γ-32P]ATP hydrolysis is determined using 0.5 μg of SWI/SNF in the absence (lanes 1-5) or presence (lanes 6-10) of nucleosomes (3 μg) and wild-type or mutant rCARM1 (0.1 μg). (D) Quantitation of SWI/SNF ATPase activity in C. The SWI/SNF activity in the presence (▪) or absence of nucleosomes (♦) is depicted.