Abstract
Background and Aims
Soil waterlogging is a major environmental stress that suppresses maize (Zea mays) growth and yield. To identify quantitative trait loci (QTL) associated with waterlogging tolerance at the maize seedling stage, a F2 population consisting of 288 F2:3 lines was created from a cross between two maize genotypes, ‘HZ32’ (waterlogging-tolerant) and ‘K12’ (waterlogging-sensitive).
Methods
The F2 population was genotyped and a base-map of 1710·5 cM length was constructed with an average marker space of 11·5 cM based on 177 SSR (simple sequence repeat) markers. QTL associated with root length, root dry weight, plant height, shoot dry weight, total dry weight and waterlogging tolerance coefficient were identified via composite interval mapping (CIM) under waterlogging and control conditions in 2004 (EXP.1) and 2005 (EXP.2), respectively.
Key Results and Conclusions
Twenty-five and thirty-four QTL were detected in EXP.1 and EXP.2, respectively. The effects of each QTL were moderate, ranging from 3·9 to 37·3 %. Several major QTL determining shoot dry weight, root dry weight, total dry weight, plant height and their waterlogging tolerance coefficient each mapped on chromosomes 4 and 9. These QTL were detected consistently in both experiments. Secondary QTL influencing tolerance were also identified and located on chromosomes 1, 2, 3, 6, 7 and 10. These QTL were specific to particular traits or environments. Although the detected regions need to be mapped more precisely, the findings and QTL found in this study may provide useful information for marker-assisted selection (MAS) and further genetic studies on maize waterlogging tolerance.
Key words: Maize (Zea mays), waterlogging tolerance, genome mapping, SSR marker, QTL, epistasis effect
INTRODUCTION
Waterlogging is one of the most important constraint factors for maize production and productivity in tropical and subtropical regions. In South-East Asia, 15 % of all maize-growing areas are affected by waterlogging problems, causing losses in maize production of 25–30 % almost every year (Rathore and Warsi, 1998). In China, the most common situation is that maize yield is limited by lack of available water, but there are large areas that are subject to waterlogging at the maize seedling stage, which is one of the most serious constraints for maize productivity, especially in south-eastern China. In these areas, high spring rainfall over a short period can lead to waterlogging of the soil for periods extending to weeks. This often causes severe damage to maize seedlings due to their poor adaptation to waterlogging. Waterlogging is becoming a matter of worldwide concern in many agricultural areas, where similar conditions have led to the spread of this environmental threat (Ghassemi et al., 1995). Predictions are that global warming will result in more erratic weather patterns, which could further exaggerate the problem. In order to increase maize productivity in waterlogged soils, new maize varieties with greater adaptation to waterlogging are essential. Hence, the development of waterlogging-tolerant varieties with a high yield potential is one of the main objectives of many maize breeding programs in the region (Anon., 2003; Zaidi et al., 2004).
The degree of stress in waterlogged soils is associated with growth stage, duration of flooding, soil type, soil acidity/alkalinity, climatic factors, growth conditions and genotypes (Rathore and Warsi, 1998; Mano et al., 2002). In a previous study (Zaidi et al., 2004), the early stages of maize development were shown to be the most sensitive to waterlogging, especially from the second leaf stage (V2) to the seventh leaf stage (V7), and roots are the first to be affected under waterlogged conditions. When the waterlogging treatment was continued for 6 d, most roots except for some adventitious ones were found to be decomposing, and plants were unable to take up the required atmospheric and edaphic nutrients, resulting in leaching and denitrification as a result of nitrogen deficiency. The latter is observed as a yellowing of the older leaves. Nitrogen deficiency itself then further increases plant stress. During waterlogging, gas exchange between soil and air decreases as gas diffusion in water is decreased 104-fold (Armstrong, 1979; Armstrong and Drew, 2002), O2 in the soil is rapidly depleted, and the soil may become hypoxic or anoxic within a few hours (Gambrell and Patrick, 1978; Malik et al., 2002). The anaerobic response of maize has been extensively reviewed previously (Sachs, 1993, 1994; Sachs et al., 1996; Mustroph and Albrecht, 2003). The inability of maize to withstand low oxygen conditions in the root zone results in substantial yield losses (Dennis et al., 2000).
In recent years, more and more information has been accumulated on the molecular, biochemical, physiological, morphological, anatomical and metabolic responses to waterlogging and oxygen deficiency in plants (Kennedy et al., 1992; Vartapetian and Jackson, 1997; Baxter-Burrell et al., 2003; Greenway et al., 2006; Mustroph et al., 2006). Tolerance to waterlogging has been studied in other crops such as wheat, soybean, Arabidopsis and rice, where it appears to be oligogenically inherited (Setter et al., 1997; Boru et al., 2001; VanToai et al., 2001; Kolk et al., 2002; Xu et al., 2006). It is interesting to note that ethylene has been reported as being involved in gene regulation after prolonged oxygen deprivation in maize (He et al., 1996), rice (Fukao et al., 2006; Xu et al., 2006) and Arabidopsis thaliana (Peng et al., 2001; Baxter-Burrell et al., 2003; McGrath et al., 2005). Saab and Sachs (1996) reported that an ethylene-signalling pathway is required for the induction of the xyloglucan endo-transglycosylase gene and that an ethylene-independent pathway is mainly responsible for the induction of ADH1 in flooded maize roots. Programmed cell death and aerenchyma formation in maize roots is regulated by ethylene, which is one important adaptation mechanism to tolerate a low-oxygen soil environment (Drew et al., 1979; Justin and Armstrong, 1991; Drew et al., 2000). In addition, Xu et al. (2006) reported that the submergence tolerance gene, Sub1A, is an ethylene-response-factor-like gene that confers submergence tolerance to rice. In Arabidopsis thaliana, ethylene was also involved in the hypoxic induction of the ADH gene (Peng et al., 2001).
Some recent studies have documented variation in the anaerobic response of maize to flooding (Sachs et al., 1996) and several morphological responses during waterlogging have been also reported (Subbaiah and Sachs, 2003). According to previous studies, the inheritance and expression of traits associated with waterlogging tolerance in maize seedlings are physiologically and genetically complex (Sachs, 1993, 1994; Liao and Lin, 2001; Subbaiah and Sachs, 2003). Complicated responses to waterlogging, such as anaerobic proteins synthesis, alterations of gene expression, metabolic (switch to a fermentative pathway) and structural changes (e.g. aerenchyma formation) have been observed. There appears to be inherent genetic variability in maize with regard to waterlogging tolerance (Sachs et al., 1996). However, manipulating waterlogging tolerance in maize is still hampered by inadequate knowledge of the molecular and physiological basis of the process.
Progress in developing improved waterlogging-tolerant cultivars would be accelerated if the genes controlling the various underlying processes could be identified and tagged with molecular markers. The goals of the current study were to create a genetic map of the traits associated with waterlogging tolerance, and to identify the QTL controlling tolerance to waterlogging at the seedling stage under controlled conditions by using segregating F2 populations derived from a cross between a waterlogging-tolerant accession (‘HZ32’) and a susceptible accession (‘K12’). The QTL analysis of physiological traits and the identification of potential candidate genes may help in understanding the genetic mechanisms of waterlogging tolerance, as well as in developing waterlogging-tolerant elite maize lines through molecular marker-assisted selection.
MATERIALS AND METHODS
Plant material and population development
An F2 mapping population was developed from a cross between two maize inbred lines, ‘HZ32’ (highly waterlogging-tolerant) and ‘K12’ ( highly waterlogging-sensitive) (Zhang et al., 2003; Tang et al., 2005). These lines had previously been identified (unpubl. res.) using the waterlogging tolerance coefficient (WTC = waterlogging treatment of each trait/control of each trait; described in detail below). Three hundred and forty-one F2 seeds derived from a single F1 parent were planted and 288 of the subsequent F2 plants were successfully self-pollinated at the experimental farm of Huazhong Agricultural University. The seeds of the 288 F2:3 ears (families) were harvested from the F2 selfed-plants in the 2003 maize-growing season. The F2 plants were used for genotyping SSR (simple sequence repeat) loci, and the F2:3 seeds (families) harvested from each F2 plants were utilized to conduct the waterlogging experiments.
Plant growth conditions
To avoid the influence of rainfall, the experiment was carried out under glasshouse conditions. The day/night temperatures were 30/22 °C, relative humidity was 55–75 % and the photoperiod was 13/11 h (day/night).
Two experiments, EXP.1 and EXP.2, were conducted in the 2004 and 2005 maize-seedling growing seasons, respectively. The same F2:3 families together with the two parents and the F1 hybrid were exposed to control (no flooding) and waterlogging treatments. The experiments were laid out in a randomized complete-block design with three replications. Two plastic pots were included for each replication per genotype: one pot for the control and the other for the waterlogging treatment. Fifteen seedlings were grown in each pot of 32 cm diameter and 32 cm depth filled with 10 kg of sieved, sterilized dry field soil (the basic physical–chemical properties of the selected soil were the same as those of the cultivated fields of the Huazhong Agricultural University) amended with 1·0 g (NH4)2SO4, 0·8 g P2O5, and 0·6 g K2O per kilogram of soil until the second leaf had fully expanded (V2 stage). The pots intended for the waterlogging treatment were then filled with water to 2–3 cm above the soil surface for 6 d. The controls were irrigated as needed to avoid drought stress or waterlogging stress.
Sampling, drying and weighing methods
After undergoing the waterlogging treatment for 6 d in both experiments, plant height (PH), root length (RL), root dry weight (RDW) and shoot dry weight (SDW) of each replicate per genotype under control and waterlogging treatment were measured. The measurements were considered to represent the phenotypes of the F2 plants.
Fifteen plants of each genotype per replicate were used for trait scoring under control and waterlogged conditions. Plants were carefully taken out of the pot and immersed in water. Roots were gently washed under running water; root loss during cleaning was kept to a minimum. Root length was measured from the coleoptilar node to the tip of the longest root and plant height was measured from the coleoptilar node to the tip of the longest leaf. Roots were then seperated from the plant. Roots and shoots of each replicate per genotype for control and waterlogging stress were put into separate paper bags, which were then rapidly transferred into ovens and dried at 65 °C until a constant weight was achieved. Root dry weight and shoot dry weight were measured using an electronic balance (MP500B, Ashiba). The waterlogging tolerance coefficient (WTC) was calculated for each pair of pots grown at the same time, with the averaged values for the fifteen plants of each pot being used. The WTC of RL, RDW, PH, SDW and total dry weight (TDW; TDW = SDW + RDW) of each replicate per genotype was calculated using the following formula:
Because the control and waterlogging treatment plants were paired for each genotype, we obtained 288 WTC values for each trait.
DNA isolation and SSR analysis
Genomic DNA from each of the F2 plants and the parental lines was isolated from fresh leaf tissue following a procedure similar to that used by Saghai-Maroof et al. (1984). The modifications in the procedure were (1) addition of boiled CTAB extraction buffer to the 50 mL polypropylene centrifuge tube, and (2) a reduction of the incubation time to 30 min.
Genotyping of the F2 individuals was performed with 177 SSR markers. Sequences of all SSR markers were obtained from the MaizeGDB database (http://www.maizegdb.org/ssr.php). Each amplification reaction contained 20 µL, consisting of 1 × reaction buffer, 10 % Glycerol, 2 mmol of MgCl2, 150 µmol of each dNTP mix, 0·3 µmol of each SSR primer, 0·75 U of Taq DNA polymerase, and 50 ng of genomic DNA. The reaction mixture was overlaid with one drop of mineral oil. Amplifications were performed in a PTC-100 Programmable Thermal Controller (MJ Research, Inc., Watertown, MA, USA) and T1 Thermocycler Module 96 (Biometro, Goettingen, Germany) programmed for the first denaturation step to last 2 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 40 s at 58 °C, and 50 s at 72 °C, with a final extension for 5 min at 72 °C. The amplified fragments were separated on 6 % polyacrylamide sequence gel containing 7 mol urea and visualized using the following silver-staining procedure. The polyacrylamide gel was fixed twice in 10 % ethanol + 0·5 % glacial acetic acid for 6 min, or fixed once for 12 min, then rinsed with ddH2O for 6 min, dipped in 0·2 % AgNO3 for 12 min of staining, rinsed with ddH2O for about 12 s, and placed in 1·5 % NaOH + 0·4 % formaldehyde (37 %) until DNA bands were displayed clearly. Later the gel was placed in 0·75 % Na2CO3 for 3 min to end the staining, and finally rinsed with tap water for about 3 min and then air-dried.
Linkage analysis and map construction
A molecular linkage map was constructed using Mapmaker Version 3·0 (Lander et al., 1987; Lincoln et al., 1993). All the markers were assigned at LOD ≥ 2·5 to ten linkage groups. By means of the Kosambi mapping function (Kosambi, 1944), the values of recombination fractions were converted into genetic map distances (cM). Linkage groups were determined using the ‘group’ command; the order of the markers for each linkage group was determined using the command ‘first order’. Ungrouped/unlinked markers were assigned to the respective linkage groups using the ‘try’ command. The map was drawn according to Liu and Meng (2003).
Statistical analysis
All the QTL analyses for the individual environments were performed using Windows QTL Cartographer Version 2·0 (North Carolina State University, Raleigh, NC) programmed by Wang et al. (2002). Composite interval mapping (CIM) was used to map the QTLs. The parameters were set as follows. Map function: Kosambi; distance units: centimorgan (cM); distance type: position; cross-information: SF3 (self-cross F3); walk speed: 2 cM; LR (likelihood ratio) threshold: 11·50 under H0:H3 (H0: a = 0, d = 0; H3: a < > 0, d < > 0), i.e., LOD = 2·5; CIM mode selection: model 6, i.e., standard model; background controls: 5 of control marker numbers and 10·0 cM of window size. Significance thresholds were determined by permutation tests (n = 1000 permutations; Churchill and Doerge, 1994); considering a significant and a suggestive locus when the LOD statistic exceeded the 95th (P < 0·05) and the 63rd (P < 0·37) percentile of the permutation distribution, respectively (Wittenburg et al., 2002; Rodrigo et al., 2006). QTL × Environment (Q × E) interaction and digenic epistatic QTLs analysis were conducted by using QTLMapper V2·0 based on a mixed model approach (Wang et al., 1999). P ≤ 0·005 for Type-I errors and a log10 likelihood ratio (LOD) value of 2·5 were used as criteria to declare the putative main effect QTL position, digenic epistatic QTLs and QTL × Environment (Q × E) interaction. Epistasis effect was estimated according to the definition of Mather and Jinks (1982). The R2 value (coefficient of determination) from this analysis indicated the percentage of phenotypic variance explained by the marker genotypes at the locus.
The PROC MIXED procedure of SAS ver. 8·02 (SAS Institute Inc., Cary, North Carolina, 1991–2001) was used to calculated the adjusted means and the broad-sense heritability (h2) of the families. The heritability was computed as h2 = δg2/(δg2 + δe2/n), where δg2 and δe2 were the estimates of genetic and residual variances, respectively, derived from the expected mean-squares of the analysis of variance, and n was the number of replications. Analysis of variance was done using the general linear model (GLM) procedure of the SAS program. The frequency distribution of the F2:3 famililies for all traits was performed using the univariate procedure of SAS and normal distributions were checked using the Shapiro–Wilk Test. Simple Pearson correlation coefficients (r) were calculated between the traits using the adjusted means of the F2:3 families. The significance of the correlation coefficient at P ≤ 0·05, 0·01, and 0·001 are indicated as *, ** and ***, respectively.
RESULTS
Phenotypic variation and phenotypic data
The frequency distribution of the F2:3 families for five waterlogging responsive traits and their WTC were normal, as determined by the Shapiro–Wilk test (data not shown). Values of the mean and range for the five traits are shown in Table 1. The parents showed statistically significant differences for the traits RL, RDW, PH, SDW and TDW under waterlogging conditions, but there were no differences under normal conditions. As shown in Table 1, the five responsive traits of all the genotypes were significantly reduced (5–58 %) in the waterlogged conditions compared with the controls in both experiments. In other words, the WTC for the five waterlogging response traits were greater for ‘HZ32’ than for ‘K12’, which indicated that the traits were more greatly affected by waterlogging in ‘K12’ than in ‘HZ32’. The results suggest that ‘HZ32’ exhibited consistently and significantly superior waterlogging tolerance for the waterlogging response traits. Moreover, the effects of waterlogging were much more severe on RL and RDW than on PH and SDW (Table 1).
Table 1.
Trait mean values for 288 F2:3 families and two parents along with broad-sense heritability (h2) in the two experiments
| Traits | Root length (cm) | Root dry weight (g) | Plant height (cm) | Shoot dry weight (g) | Total dry weight (g) |
|---|---|---|---|---|---|
| EXP.1 (2004) | |||||
| Control | |||||
| ‘HZ32’ (P1) | 33·66 ± 1·21 | 0·16 ± 0·02 | 28·43 ± 1·62 | 0·41 ± 0·01 | 0·57 ± 0·02 |
| ‘K12’ (P2) | 32·56 ± 1·21 | 0·12 ± 0·02 | 24·23 ± 1·62 | 0·38 ± 0·01 | 0·50 ± 0·02 |
| P1 vs P2† | ns | ns | ns | ns | ns |
| F2:3 families (mean) | 33·95 ± 2·62 | 0·17 ± 0·02 | 24·52 ± 1·31 | 0·24 ± 0·01 | 0·41 ± 0·03 |
| F2:3 families (range) | 11·7–47·2 | 0·04–0·48 | 13·5–35·5 | 0·08–0·57 | 0·14–1·05 |
| h2 | 0·89 | 0·86 | 0·82 | 0·78 | 0·84 |
| Waterlogging stress | |||||
| ‘HZ32’ (P1) stress | 19·40 ± 1·40 | 0·11 ± 0·01 | 27·19 ± 3·21 | 0·35 ± 0·02 | 0·46 ± 0·02 |
| ‘K12’ (P2) stress | 14·95 ± 1·40 | 0·05 ± 0·01 | 20·5 ± 3·21 | 0·19 ± 0·02 | 0·24 ± 0·02 |
| P1 vs P2 | * | ** | ** | ** | ** |
| F2:3 families (mean) | 16·7 ± 1·34 | 0·06 ± 0·01 | 21·10 ± 1·96 | 0·20 ± 0·03 | 0·28 ± 0·01 |
| F2:3 families (range) | 3·9–34·25 | 0·01–0·24 | 8·96–31·06 | 0·04–0·39 | 0·06–0·96 |
| h2 | 0·28 | 0·43 | 0·74 | 0·71 | 0·82 |
| WTC of P1 | 0·58 | 0·68 | 0·95 | 0·85 | 0·81 |
| WTC of P2 | 0·46 | 0·42 | 0·85 | 0·50 | 0·48 |
| EXP.2 (2005) | |||||
| Control | |||||
| ‘HZ32’ (P1) | 26·32 ± 1·41 | 0·23 ± 0·01 | 36·81 ± 1·13 | 0·77 ± 0·02 | 1·00 ± 0·01 |
| ‘K12’ (P2) | 29·76 ± 1·41 | 0·17 ± 0·01 | 33·72 ± 1·13 | 0·69 ± 0·02 | 0·86 ± 0·01 |
| P1 vs P2 | ns | ns | ns | ns | ns |
| F2:3 families (mean) | 39·1 ± 1·52 | 0·42 ± 0·01 | 35·39 ± 1·92 | 0·59 ± 0·02 | 1·03 ± 0·01 |
| F2:3 families (range) | 26–55·2 | 0·15–0·83 | 24·1–44·94 | 0·14–1·2 | 0·42–2·03 |
| h2 | 0·72 | 0·83 | 0·51 | 0·62 | 0·67 |
| Waterlogging stress | |||||
| ‘HZ32’ (P1) | 19·12 ± 1·23 | 0·16 ± 0·01 | 26·84 ± 1·62 | 0·62 ± 0·02 | 0·78 ± 0·03 |
| ‘K12’ (P2) | 14·85 ± 1·23 | 0·09 ± 0·01 | 18·30 ± 1·62 | 0·51 ± 0·02 | 0·60 ± 0·02 |
| P1 vs P2 | * | ** | ** | ** | ** |
| F2:3 families (mean) | 14·69 ± 1·30 | 0·14 ± 0·02 | 25·96 ± 1·54 | 0·39 ± 0·01 | 0·54 ± 0·01 |
| F2:3 families (range) | 5·8–22·5 | 0·03–0·53 | 15·3–40·1 | 0·14–0·8 | 0·1–1·08 |
| h2 | 0·32 | 0·57 | 0·69 | 0·58 | 0·61 |
| WTC of P1 | 0·73 | 0·70 | 0·73 | 0·80 | 0·78 |
| WTC of P2 | 0·50 | 0·52 | 0·54 | 0·73 | 0·70 |
The heritability was computed as h2 = δ2g/(δ2g + δ2e/n), where δ2g and δ2e were the estimates of genetic and residual variances, respectively, derived from the expected mean-squares of the analysis of variance, and n was the number of replications. WTC (waterlogging tolerance coefficient) was computed as WTC = waterlogging treatment of each trait/control of each trait. Data are mean ± s.e.m.
† Statistical test for difference between two parents at 0·05 (*) and 0·01 (**) levels of probability; ns, not significant.
The F2:3 families exhibited a wide range of variation for the traits studied (Table 1). Transgressive segregation in both directions was observed for most traits under waterlogging conditions, indicating that both parents transmitted favourable alleles for each trait. Broad-sense heritabilities (h2) computed across the five traits mentioned were relatively moderate (Table 1). PH and TDW had the highest h2 at 0·74 and 0·82, while RL had the lowest at 0·28 and 0·32 in EXP.1 and EXP.2, respectively, under waterlogging-stress conditions. Comparing with h2 under control conditions, the results indicated that RL was more prone to be affected by waterlogging stress than other traits (Table 1).
Correlations between measured traits and WTC were evaluated for statistical significance in both experiments as shown in Tables 2 and 3. Highly significant positive correlations between TDW and PH, TDW and SDW, TDW and RDW, and RL and RDW were found in both experiments. Positive correlations between PH and SDW, and PH and RDW were also highly significant in EXP.2 (Table 2). In addition, highly significant positive correlations among WTC of the five response traits assessed were found in both two experiments (Table 3), indicating that these traits were not expressed independently of one another. Highly significant correlations were observed between EXP.1 and EXP.2 for TDW, PH, SDW and their WTC (Tables 2 and 3). However, a weak relationship was observed for RDW (r = 0·4786) and no relationship was found for RL between the two experiments (Table 2). Similar results were found for WTC of RDW and RL (Table 3), suggesting that the expression of RDW and RL is sensitive to the growing environment.
Table 2.
Simple correlation coefficients between waterlogging-response traits in maize obtained in a F2 mapping population derived from a cross between waterlogging-tolerant ‘HZ32’ and waterlogging-sensitive ‘K12’ in EXP.1 (2004 season) and EXP.2 (2005 season)
| EXP.2 (2005) | |||||
|---|---|---|---|---|---|
| Total dry weight | Plant height | Root length | Shoot dry weight | Root dry weight | |
| EXP.1 (2004) | |||||
| Total dry weight | 0·8456*** | 0·7163*** | 0·3430** | 0·8768*** | 0·8925*** |
| Plant height | 0·74826*** | 0·6785*** | 0·3854* | 0·6360*** | 0·5609*** |
| Root length | 0·31448** | 0·3401* | 0·2345ns | 0·2782* | 0·6788*** |
| Shoot dry weight | 0·8631*** | 0·7327** | 0·3723** | 0·7843*** | 0·2863* |
| Root dry weight | 0·8358*** | 0·27291** | 0·7843*** | 0·6538** | 0·4786*** |
Values in italics denote a correlation between the two experiments.
*, significant at P < 0·05; **, P < 0·01; ***, P < 0·001; ns, not significant.
Table 3.
Simple correlation coefficients between the waterlogging tolerance coefficient (WTC = waterlogging treatment of each trait/control of each trait) for five waterlogging-response traits in maize obtained in a F2 mapping population derived from a cross between waterlogging-tolerant ‘HZ32’ and waterlogging-sensitive ‘K12’ in EXP.1 (2004 season) and EXP.2 (2005 season)
| EXP.2 (2005) | |||||
|---|---|---|---|---|---|
| Total dry weight | Plant height | Root length | Shoot dry weight | Root dry weight | |
| EXP.1 (2004) | |||||
| Total dry weight | 0·836*** | 0·72073*** | 0·34648*** | 0·89889*** | 0·72193*** |
| Plant height | 0·64826*** | 0·683*** | 0·3783*** | 0·70882*** | 0·45982*** |
| Root length | 0·51448*** | 0·64001*** | 0·345ns | 0·30955*** | 0·27884*** |
| Shoot dry weight | 0·66317*** | 0·52703*** | 0·35719*** | 0·820*** | 0·3863*** |
| Root dry weight | 0·72258*** | 0·47291*** | 0·4111*** | 0·69853*** | 0·453*** |
Values in italics denote a correlation between the two experiments.
***, significant at P < 0·001; ns, not significant.
Construction of molecular marker linkage map
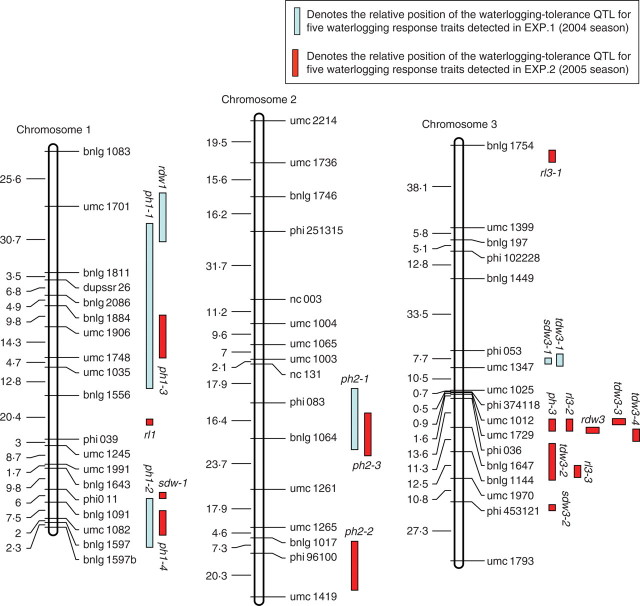
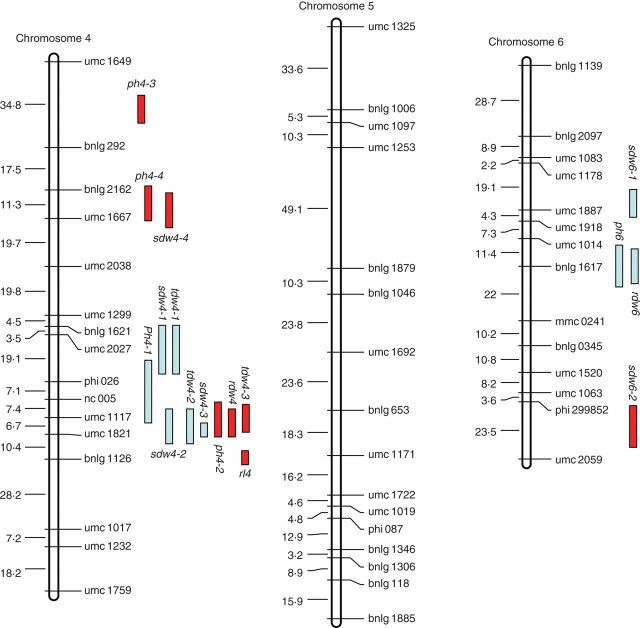
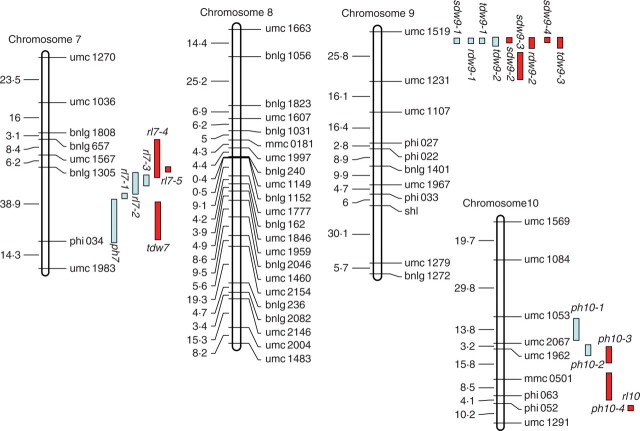
One hundred and seventy-seven SSR markers showing co-dominant segregation were employed to construct a linkage map (Fig. 1), of which 148 informative markers were assigned to ten chromosomes based on LOD values exceeding 11·5. The linkage map had a total length of 1710·5 cM with an average interval of 11·5 cM between adjacent markers.
Fig. 1.
Molecular linkage map of the F2 population derived from a cross between ‘HZ32’ and ‘K12’, and summary of QTL for all traits responsive to waterlogging in the mapping population of maize in EXP.1 and EXP.2. sdw = shoot dry weight; ph = plant height; rl = root length; rdw = root dry weight; tdw = total dry weight. For all the QTL names, the first number following the letters represents the chromosome locations of the QTL and the second number represents the orders of the QTL located on the same chromosome by the same trait. The distances between markers (cM) are listed to the left of each figure part.
QTL detection for PH, SDW, RL, RDW and TDW
QTL for traits responsive to waterlogging in both experiments mapped to maize chromosomes 1, 2, 3, 4, 6, 7, 9 and 10 (Tables 4 and 5, Fig. 1). A total of 59 putative QTL were found to be associated with the five waterlogging-response traits and their WTC when the results of the two experiments were considered together. Fourteen and 25 QTL were detected under control and waterlogging treatment conditions, respectively. Twenty QTL were identified for WTC (Tables 4 and 5). The detected QTL individually accounted for 3·9–37·3 % of the phenotypic variation. Out of the total of 59, 13 QTL individually accounted for more than 10 % of the phenotypic variation. A list of the putative QTL flanked by SSR markers along with their phenotypic variance, additive effects and peak LOD scores, are presented in Tables 4 and 5. A graphical presentation of QTL locations on the linkage map is shown in Fig. 1.
Table 4.
Map position and main characteristics of QTL with a LOD score ≥2·5 for plant height (ph), shoot dry weight (sdw), root length (rl), root dry weight (rdw), total dry weight (tdw = sdw + rdw) and waterlogging tolerance coefficient (WTC = waterlogging treatment of each trait/control of each trait) detected in a F2 population of maize derived from a cross between waterlogging-tolerant ‘HZ32’ and waterlogging-sensitive ‘K12’ in EXP.1 (2004 season)
| Traits | QTLa | Chromosome number | cMb | Rangec | Nearest marker | LODd | R2 ( %)e | Additivityf |
|---|---|---|---|---|---|---|---|---|
| Control | ||||||||
| Plant height | ph1-1 | 1 | 75 | 27–109 | bnlg1556 | 9·04 | 13·5 | 1·374 |
| ph1-2 | 1 | 166 | 160–174 | bnlg1643 | 3·83 | 6·0 | −1·459 | |
| ph4-1 | 4 | 141 | 136–149 | umc2027 | 3·03 | 4·7 | 0·743 | |
| ph7 | 7 | 98 | 85–105 | umc1567 | 3·06 | 4·9 | 1·552 | |
| Shoot dry weight | sdw4-1 | 4 | 111 | 109–118 | umc1299 | 2·73 | 4·2 | 2·196 |
| Root length | rl7-1 | 7 | 79 | 77–82 | umc1567 | 2·53 | 7·4 | 1·892 |
| Root dry weight | – | – | – | – | – | – | – | – |
| Total dry weight | tdw4-1 | 4 | 111 | 109–118 | umc1299 | 3·02 | 4·6 | 0·319 |
| Waterlogging treatment | ||||||||
| Plant height | ph2-1 | 2 | 147 | 140–154 | nc131 | 2·86 | 4·2 | −0·054 |
| ph6 | 6 | 81 | 76–90 | umc1918 | 3·85 | 5·8 | 1·831 | |
| Shoot dry weight | sdw3-1 | 3 | 95 | 95 | umc1347 | 2·50 | 3·9 | −0·647 |
| sdw4-2 | 4 | 148 | 143–158 | phi026 | 3·34 | 5·6 | −1·747 | |
| sdw9-1 | 9 | 2·0 | 2·0 | umc1519 | 15·19 | 37·3 | 0·385 | |
| Root length | rl7-2 | 7 | 61 | 53–76 | bnlg657 | 3·33 | 6·3 | −2·243 |
| Root dry weight | rdw6 | 6 | 80 | 78–82 | umc1918 | 2·56 | 4·2 | 0·978 |
| rdw9-1 | 9 | 2·0 | 2·0 | umc1519 | 3·29 | 26·3 | 0·366 | |
| Total dry weight | tdw3-1 | 3 | 95 | 93–96 | umc1347 | 2·63 | 4·1 | -0·073 |
| tdw4-2 | 4 | 148 | 143–158 | phi026 | 3·21 | 5·5 | -0·223 | |
| tdw9-1 | 9 | 2·0 | 2·0 | umc1519 | 10·17 | 33·3 | 0·072 | |
| WTC | ||||||||
| Plant height | ph10-1 | 10 | 57 | 53–62 | umc2067 | 3·06 | 7·1 | −0·061 |
| ph10-2 | 10 | 66 | 65–71 | umc1053 | 3·23 | 5·2 | −0·062 | |
| Shoot dry weight | sdw4-3 | 4 | 150 | 150–152 | nc005 | 2·60 | 5·1 | −0·104 |
| sdw6-1 | 6 | 58 | 49–59 | umc1918 | 2·96 | 5·4 | 0·024 | |
| Root length | rl7-3 | 7 | 57 | 55–62 | bnlg657 | 2·68 | 4·0 | −0·060 |
| Root dry weight | rdw1 | 1 | 25 | 18–34 | bnlg1811 | 2·89 | 4·4 | −0·627 |
| Total dry weight | tdw9-2 | 9 | 2·0 | 2·0–4·0 | umc1519 | 5·92 | 31·7 | 0·045 |
aThe first number following the letters represents the chromosome locations of the QTL and the second number represents the orders of the QTL located on the same chromosome by the same trait.
bPosition of the peak of the QTL in centimorgans.
cRange of the QTL above the threshold LOD score.
dLOD score calculated by WinQTLCart 2·0.
ePercentage of the phenotypic variance explained by genotype class at QTL peak.
fAdditivity: positive additivity indicates that the high values of the trait were inherited from the tolerant parent (‘HZ32’); negative additivity indicatess that the high values of the trait were inherited from the sensitive parent (‘K12’).
Table 5.
Map position and main characteristics of QTLs with a LOD score ≥2·5 for plant height (ph), shoot dry weight (sdw), root length (rl), root dry weight (rdw), total dry weight (tdw = sdw + rdw) and waterlogging tolerance coefficient (WTC = waterlogging treatment of each trait/control of each trait) detected in a F2 population of maize derived from a cross between waterlogging-tolerant ‘HZ32’ and waterlogging-sensitive ‘K12’ in EXP.2 (2005 season)
| Traits | QTLa | Chromosome number | cMb | Rangec | Nearest marker | LODd | R2 ( %)e | Additivityf |
|---|---|---|---|---|---|---|---|---|
| Control | ||||||||
| Plant height | ph1-3 | 1 | 89 | 68–95 | umc1748 | 4·81 | 11 | 1·977 |
| ph2-2 | 2 | 212 | 199–221 | umc1265 | 3·26 | 9·4 | −1·738 | |
| ph4-2 | 4 | 141 | 136–151 | umc1821 | 3·08 | 6·5 | −0·105 | |
| Shoot dry weight | sdw1 | 1 | 157 | 157 | bnlg1643 | 2·76 | 5·7 | −0·020 |
| sdw6-2 | 6 | 158 | 146–159 | umc1063 | 3·82 | 8·5 | 0·092 | |
| Root length | – | – | – | – | – | – | – | – |
| Root dry weight | rdw4 | 4 | 143 | 141–147 | phi026 | 2·91 | 6·5 | −0·089 |
| Total dry weight | tdw4-3 | 4 | 143 | 139–149 | phi026 | 3·24 | 7·5 | −0·320 |
| Waterlogging treatment | ||||||||
| Plant height | ph1-4 | 1 | 174 | 164–175 | phi011 | 3·74 | 7·0 | −1·140 |
| ph2-3 | 2 | 161 | 147–179 | phi083 | 5·10 | 14·3 | −1·661 | |
| ph4-3 | 4 | 18 | 16–20 | bnlg292 | 2·58 | 10·3 | 0·130 | |
| ph4-4 | 4 | 60 | 56–64 | bnlg292 | 2·76 | 6·1 | 0·239 | |
| ph10-3 | 10 | 72 | 66–79 | umc1053 | 2·67 | 5·5 | 0·496 | |
| ph10-4 | 10 | 88 | 84–91 | phi063 | 2·65 | 5·3 | 0·347 | |
| Shoot dry weight | sdw4-4 | 4 | 63 | 60–68 | bnlg292 | 3·06 | 6·3 | 0·006 |
| sdw9-2 | 9 | 2·0 | 2·0 | umc1519 | 2·68 | 7·2 | 0·029 | |
| sdw9-3 | 9 | 24 | 12–24 | umc1231 | 3·13 | 6·7 | 0·047 | |
| Root length | rl4 | 4 | 161 | 161–162 | umc1117 | 2·58 | 5·2 | 0·004 |
| rl7-4 | 7 | 55 | 50–62 | bnlg1808 | 2·64 | 5·5 | 1·737 | |
| Root dry weight | rdw9-2 | 9 | 2·0 | 2·0–4·0 | umc1519 | 6·7 | 36·3 | 2·609 |
| Total dry weight | tdw3-2 | 3 | 125 | 119–133 | umc1729 | 3·61 | 9·5 | −0·754 |
| tdw7 | 7 | 96 | 87–100 | umc1983 | 2·73 | 5·4 | 0·704 | |
| WTC | ||||||||
| Plant height | ph3 | 3 | 114 | 114–115 | umc1729 | 2·66 | 5·4 | −0·439 |
| Shoot dry weight | sdw3-2 | 3 | 154 | 154–155 | phi453121 | 2·68 | 5·6 | −0·087 |
| sdw9-4 | 9 | 0 | 0–2 | umc1519 | 7·03 | 20·8 | −0·034 | |
| Root length | rl1 | 1 | 136 | 135–137 | bnlg1556 | 2·74 | 5·1 | −0·037 |
| rl3-1 | 3 | 0 | 0–6 | umc1399 | 2·80 | 5·3 | −0·702 | |
| rl3-2 | 3 | 114 | 114–116 | umc1025 | 3·49 | 6·7 | −0·035 | |
| rl3-3 | 3 | 125 | 125–128 | bnlg1647 | 2·5 | 6·1 | −0·034 | |
| rl7-5 | 7 | 51 | 50–51 | bnlg1305 | 2·79 | 5·5 | 0·031 | |
| rl10 | 10 | 102 | 102–105 | mmc0501 | 2·77 | 5·6 | 0·027 | |
| Root dry weight | rdw3 | 3 | 117 | 117 | umc1729 | 2·67 | 5·2 | −1·955 |
| Total dry weight | tdw3-3 | 3 | 114 | 114 | phi374118 | 2·64 | 12·2 | −0·652 |
| tdw3-4 | 3 | 119 | 117–124 | umc1729 | 2·77 | 12·8 | −0·749 | |
| tdw9-3 | 9 | 2·0 | 2·0–4·0 | umc1519 | 5·91 | 30·7 | 0·065 |
aThe first number following the letters represents the chromosome locations of the QTL and the second number represents the orders of the QTLs located on the same chromosome by the same trait.
bPosition of the peak of the QTL in centimorgans.
cRange of the QTL above the threshold LOD score.
dLOD score calculated by WinQTLCart 2·0.
ePercentage of the phenotypic variance explained by genotype class at QTL peak.
fAdditivity: positive additivity indicates that the high values of the trait were inherited from the tolerance parent (‘HZ32’); negative additivity means that the high values of the trait were inherited from the sensitive parent (‘K12’).
For PH, 15 QTL were detected under control and waterlogging treatment conditions on chromosomes 1 (ph1-1, ph1-2, ph1-3 and ph1-4), 2 (ph2-1, ph2-2 and ph2-3), 4 (ph4-1, ph4-2, ph4-3 and ph4-4), 6 (ph6), 7 (ph7) and 10 (ph10-3 and ph10-4) (Tables 4 and 5). Out of these 15, six and nine QTL were detected in EXP.1 and EXP.2, respectively. Individual QTL accounted for 4·2–14·3 % of the phenotypic variation. For six of the QTL (ph1-2, ph2-1, ph2-2, ph4-2, ph1-4 and ph2-3), alleles from ‘K12’ contributed towards an increase of the trait values. For the other nine QTL, alleles from ‘HZ32’ tended to increase the trait value.
Nine QTL were mapped for SDW on chromosomes 1 (sdw1), 3 (sdw3), 4 (sdw4-1, sdw4-2, sdw4-3 and sdw4-4), 6 (sdw6-2) and 9 (sdw9-1, sdw9-2 and sdw9-3) (Tables 4 and 5). Individual QTL accounted for 3·9–37·2 % of the phenotypic variation. For six of these QTL (sdw4-1, sdw4-4, sdw6-2, sdw9-1, sdw9-2 and sdw9-3), alleles from ‘HZ32’ tended to increase the trait values, whereas for the other three QTL the allele from ‘K12’ contributed to the increase in the trait score (Tables 4 and 5).
Only one QTL for RL, rl7-1, was detected under control conditions. Three QTL (rl4, rl7-2 and rl7-4) were detected under waterlogged conditions (Tables 4 and 5). Individual QTL had values of R2 ranging from 5·2–5·5 % and 6·3–7·4 % for EXP.1 and EXP.2, respectively. Except for QTL rl7-2, all the alleles were from ‘HZ32’.
Of the four QTL associated with RDW, two (rdw9-1 and rdw9-2) were found in both experiments. Out of the remaining two QTL, one (rdw4) on chromosome 4 was detected only in EXP.2 whereas the other (rdw6) on chromosome 6 was found only in EXP.1 (Tables 4 and 5). The QTL affecting RDW, rdw9-1 and rdw9-2, explained 26·3 % and 36·3 % of the phenotypic variation in both experiments, respectively, suggesting that most of the major QTL for this trait have been identified. This finding is in good agreement with the high heritability estimates of this trait in both experiments (Table 1).
Seven genomic regions were detected to be associated with TDW. Out of the seven, two QTL, tdw4-2 and tdw4-3 on chromosome 4, were common in both experiments, indicating their low sensitivity to environmental changes, which is in good agreement with the observed phenotypic correlation between the two experiments for this trait (r = 0·8456). Out of the remaining five QTL, three (tdw3-1, tdw4-1 and tdw9-1) on chromosomes 3, 4 and 9 were detected only in EXP.1, whereas the other two (tdw3-2 and tdw7) were found only in EXP.2. Individual QTL had values of R2 ranging from 4·1–33·3 % and 5·4–9·5 % for EXP.1 and EXP.2, respectively. Four QTL had alleles from ‘K12’, the exceptions being tdw4-1, tdw7 and tdw9-1 (Tables 4 and 5).
QTL detection for WTC
A total of 20 putative QTL were found to be associated with the WTC of the five waterlogging-response traits when the results of the two experiments were considered together. The detected QTL individually accounted for 4·0–31·7 % of the phenotypic variation. Out of the 20, five QTL individually accounted for more than 10 % of the phenotypic variation (Tables 4 and 5).
Two QTL (ph10-1 and ph10-2) for the WTC of PH were detected on chromosome 10 in EXP.1. They could explain 3·06 % and 3·23 % of the total phenotypic variation and the primary effect was negative-additive, meaning that alleles from ‘K12’ at ph10-1 and ph10-2 operate in the direction of increasing the WTC of PH (Tables 4 and 5). In addition, only one QTL (ph3) for the WTC of PH was detected on chromosome 3, which could explain 5·4 % of the phenotypic variation. The ‘K12’ alleles at ph3 increased the WTC of PH.
Out of four QTL associated with the WTC of SDW, two (sdw4-3 and sdw6-1) were detected in EXP.1 and the other two (sdw3-2 and sdw9-4) were detected in EXP.2. No common QTL were found between the two seasons (Tables 4 and 5).
Seven QTL were detected for the WTC of RL on chromosomes 1, 3, 7 and 10 in the two experiments (Tables 4 and 5). The QTL rl7-3 was detected on chromosome 7 and explained 4·0 % of the phenotypic variation in EXP.1. In EXP.2, six QTL (rl1, rl3-1, rl3-2, rl3-3, rl7-5 and rl10) individually had R2 values ranging from 5·1 to 6·7 %. No common QTL were found between the two seasons. Except for rl7-5 and rl10, all alleles were from ‘K12’.
Only two QTL (rdw1 and rdw3) were detected for the WTC of RDW in the two experiments. One (rdw1) was detected in EXP.1 while the other (rdw3) was detected in EXP.2, They only explained 4·4 % and 5·2 % of the total phenotypic variation and the primary effect was negative-additive, meaning that alleles from ‘K12’ at rdw1 and rdw3 operate in the direction of increasing the WTC of RDW (Tables 4 and 5).
Four QTL were detected to be associated with TDW. Out of the four, two (tdw9-2 and tdw9-3) were common in both experiments. They could explain 31·7 % and 30·7 % of the total phenotypic variation and the primary effect was additive, meaning that alleles from ‘HZ32’ at tdw9-2 and tdw9-3 operate in the direction of increasing the WTC of TDW (Tables 4 and 5). The other two QTL (tdw3-3 and tdw3-4) located on chromosome 3 were detected in EXP.2. They could explain 12·2 % and 12·8 % of the phenotypic variation. The ‘K12’ alleles at tdw3-3 and tdw3-4 increased the WTC of TDW.
QTL groups
Taken together, only six QTL (rl1, ph2-2, rl3-1, ph4-3, sdw6-1 and sdw6-2) were not mapped close to other QTL (Fig. 1). The remaining 53 QTL were detected in the same chromosome regions, forming 11 groups. The highest concentration of QTL was found in the umc1519–umc1231 marker interval on chromosome 9. Other important groups that were found were on chromosomes 3, 4 and 7, where QTL for more than two traits each were detected (Fig. 1). The results indicate that these regions are under related genetic control.
Epistatic effect
Significant epistatic loci (P ≤ 0·005) for WTC and all target traits under control and waterlogging-stress conditions were detected by the QTLmapper (Wang et al., 1999; Table 6). Means of the results from 2004 and 2005 were used as input data for the analysis. In control conditions, a total of eight epistatic interactions were detected for all traits. One significant QTL pair for each trait was detected in sdw, rl and tdw. The epistatic effect (AAij) explained 6·78 % to 7·46 % of total variance. Five QTL pairs were detected for ph, contributing from 4·62 % to 11·81 % of the variance. Three pairs of epistatic loci involved two intervals, both having a significant putative QT locus while the other five QTL pairs involved one putative QT locus. No espistatic QT locus was detected for rdw. In waterlogging-stress conditions, ten epistatic QTL pairs were detected. The contribution rates of a single QTL pair varied from 7·6 to 33·47 %. Two pairs of epistatic loci in sdw and tdw involved two intervals both having a significant putative QT locus, and eight epistatic QTL involved one interval having a significant putative QT locus (Table 6). For the WTC of the used traits, ten epistatic QTL pairs were detected. The contribution rates of a single QTL pair varied from 5·98 to 25·49 %. Four pairs of epistatic loci (one for ph, one for sdw and two for rl) involved two intervals both having a significant putative QT locus, while the other six QTL pairs involved one putative QT locus.
Table 6.
Epistatic loci for plant height (PH), shoot dry weight (SDW), root length (RL), root dry weight (RDW), total dry weight (TDW = SDW + RDW) and the waterlogging tolerance coefficient (WTC = waterlogging treatment of each trait/control of each trait) under control and waterlogging-stress conditions in maize
| Trait | Chromosome interval i | Interval i | Chromosome interval j | Interval j | LODa | Abic | R2 (Ai)d | Aje | R2(Aj) | AAfij | R2 (AAij)g |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Epistasis loci detected under control | |||||||||||
| ph | 1–6 | bnlg1884-umc1906 | 4–8 | umc2027-phi026 | 9·3 | 1·97 | 4·5 | 0·4 | 5·4 | 2·09 | 8·92 |
| 1–6 | bnlg1884-umc1906 | 4–14 | umc1017-umc1232 | 8·57 | 1·96 | 13·0 | 1·43 | 11·81 | |||
| 1–7 | umc1906-umc1748 | 4–8 | umc2027-phi026 | 4·09 | 2·07 | 3·45 | 0·54 | 4·1 | 2·43 | 4·62 | |
| 2–14 | bnlg1017-phi96100 | 5–3 | umc1097-umc1253 | 4·28 | −1·43 | 6·35 | −1·44 | 7·18 | |||
| 2–1 | umc2214-umc1736 | 4–11 | umc1117-umc1821 | 6·04 | −0·89 | 10·2 | −2·22 | 9·42 | |||
| sdw | 1–14 | bnlg1643-phi011 | 4–7 | bnlg1621-umc2027 | 3·59 | −0·01 | 6·2 | 1·34 | 5·8 | −1·21 | 6·78 |
| rl | 7–6 | bnlg1305-phi034 | 8–3 | bnlg1823-umc1607 | 3·42 | 1·23 | 8·4 | 2·17 | 7·46 | ||
| tdw | 1–18 | bnlg1597-bnlg1597b | 4–8 | umc2027-phi026 | 4·95 | 0·23 | 6·52 | 0·28 | 7·12 | ||
| Epistasis loci detected under waterlogging stress | |||||||||||
| ph | 2–10 | phi083-bnlg1064 | 5–9 | umc1171-umc1722 | 7·98 | −2·1 | 8·3 | −3·44 | 11·9 | ||
| 5–15 | bnlg118-bnlg1885 | 4–3 | bnlg2162-umc1667 | 7·59 | 0·49 | 10·56 | 0·34 | 11·9 | |||
| sdw | 9–1 | umc1519-umc1231 | 7–5 | umc1567-bnlg1305 | 8·12 | 0·03 | 18·9 | −0·54 | 8·9 | −0·81 | 16·8 |
| 5–4 | umc1253-bnlg1879 | 4–11 | umc1117-umc1821 | 5·32 | −1·02 | 9·47 | 1·26 | 8·69 | |||
| 9–1 | umc1519-umc1231 | 10–1 | umc1569-umc1084 | 13·29 | 0·1 | 21·89 | 0·15 | 19·87 | |||
| rl | 7–6 | bnlg1305-phi034 | 3–2 | umc1399-bnlg197 | 3·56 | 1·56 | 8·23 | 1·87 | 7·63 | ||
| rdw | 6–7 | umc1014-bnlg1617 | 9–2 | umc1231-umc1107 | 8·66 | 0·01 | 7·78 | 0·02 | 8·16 | ||
| 9–1 | umc1519-umc1231 | 10–1 | umc1569-umc1084 | 9·69 | 0·03 | 29·6 | 0·13 | 30·3 | |||
| tdw | 4–8 | umc2027-phi026 | 4–11 | umc1117-umc1821 | 11·7 | 0·77 | 10·81 | −0·76 | 9·45 | −0·78 | 11·46 |
| 9–1 | umc1519-umc1231 | 10–4 | umc2067-umc1962 | 11·05 | 0·16 | 29·46 | 0·21 | 33·47 | |||
| Epistasis loci detected by WTC | |||||||||||
| ph | 4–11 | umc1117-umc1821 | 8–15 | bnlg2046-umc1460 | 7·91 | −0·04 | 7·64 | −0·09 | 6·98 | ||
| 4–3 | bnlg2162-umc1667 | 8–2 | bnlg1056-bnlg1823 | 5·74 | 0·35 | 7·21 | 0·37 | 7·86 | |||
| 3–10 | umc1012-umc1729 | 1–7 | umc1906-umc1748 | 4·82 | 0·02 | 9·86 | −0·07 | 5·78 | −0·12 | 8·79 | |
| 10–4 | umc2067-umc1962 | 9–4 | phi027-phi022 | 3·56 | 0·41 | 5·68 | 0·45 | 6·78 | |||
| sdw | 4–3 | bnlg2162-umc1667 | 9–1 | umc1519-umc1231 | 9·58 | 0·05 | 24·2 | 0·03 | 17·37 | 0·22 | 25·49 |
| rl | 7–5 | umc1567-bnlg1305 | 1–2 | umc1701-bnlg1811 | 4·23 | 0·32 | 6·34 | −0·61 | 5·67 | −0·82 | 7·23 |
| 1–10 | bnlg1556-phi039 | 3–10 | umc1012-umc1729 | 3·89 | −0·06 | 5·96 | −0·09 | 6·89 | −0·14 | 8·37 | |
| rdw | 3–10 | umc1012-umc1729 | 5–8 | bnlg653-umc1171 | 2·81 | −0·09 | 6·46 | −0·19 | 5·98 | ||
| tdw | 9–1 | umc1519-umc1231 | 10–8 | phi052-umc1291 | 8·7 | 0·01 | 19·8 | 0·02 | 17·8 | ||
| 2–4 | phi251315-nc003 | 9–1 | umc1519-umc1231 | 11·7 | 0·38 | 19·8 | 0·97 | 20·6 |
The means of 2004 and 2005 were used as input data.
aLOD score calculated by QTLmapper 2·0 at P ≤ 0·005 level of probability.
bAdditive effect.
cThe estimates of additive effect for testing point i.
dPercentage of the phenotypic variance explained by the marker genotypes at the locus.
eAdditive effect for testing point j.
fAdditive × additive epistasis.
gAdditive by additive effects between the two testing points i and j.
DISCUSSION
Phenotypic variation
Traits of the two parental lines responsive to waterlogging differed considerably when the plants were exposed to waterlogging-stress conditions. These differences disappeared when the plants developed under control conditions. The data in Table 1 suggest that ‘K12’ was more severely affected by waterlogging whilst ‘HZ32’ was more tolerant to waterlogging. In other words, higher values for the traits investigated indicated higher waterlogging tolerance. Since the parental lines were selected on the basis of high or low WTC under artificial control conditions during the maize seedling growing season in our previous study (unpubl. data), this observation indicates that the selection method was efficient in this respect. Moreover, the fact that the favourable alleles for most of the identified QTL were inherited from the waterlogging-tolerant parent also shows the efficacy of the selection method.
The strong phenotypic correlations among traits responsive to waterlogging stress that were observed in the two parental lines and the F2:3 families indicated common mechanisms for waterlogging tolerance (Tables 2 and 3). Classical quantitative genetics assumes that trait correlation is a causal effect of pleiotropy or an effect of closely linked genes. Therefore, it would be expected that the QTL for the correlated traits would be mapped in similar genomic regions. In the present study, PH and SDW possess three common genomic regions in chromosomes 1 and 4 (Fig. 1). These morphological characters were also mapped in regions very close to the QTL for TDW in chromosomes 3, 4, 7 and 9 (Fig. 1). PH, SDW and TDW were significantly positive correlated (Tables 2 and 3), which is in agreement with the observations as the QTLs of these characters were mapped in genomically similar regions (Fig. 1). The positive relationships between the other responsive traits also support this hypothesis (Tables 2 and 3). This finding is in line with the opinions expressed by Materechera et al. (1992) and Ali et al. (2000).
Significant variation and a normal distribution for the traits studied made this population suitable for QTL analysis. Except for RL in EXP.2, the mean values of the population were close to the mid-parental values for all traits in both experiments (Table 1). Although the phenotypic data for F2:3 were distributed normally, transgressive segregation was observed in both directions for all traits, indicating that neither of the parents carried all the positive or all negative alleles.
A major QTL for waterlogging tolerance is located on chromosome 9
Major QTL controlling traits associated with SDW, RDW, TDW, the WTC of TDW and the WTC of SDW all mapped to the same region of chromosome 9 and were consistently identified in the both experiments. The expression of waterlogging tolerance is known to be environmentally dependent and genetically complex (Sachs, 1993, 1994; Liao and Lin, 2001; Subbaiah and Sachs, 2003). For other crops, such as rice, in different years and seasons and with different mapping populations, the QTL controlling traits related to waterlogging tolerance have been mapped on many genomic regions (Xu and Mackill, 1996; Toojinda et al., 2003). However, the consistently detected major QTL indicated that this region on chromosome 9 is important in the waterlogging response in this maize population; indeed, the most important waterlogging-tolerance QTL in this study. Moreover, these QTL were only for dry matter accumulation and were not associated with root length and plant height. The QTL were only detected under waterlogging-stress conditions in both experiments, so we presume that there is a specific waterlogging-tolerance responsive gene. It is worthwhile considering the association between the identified QTL controlling waterlogging tolerance and genes known to be regulated by anoxia, which provides us with some genetic evidence that some genes responsive to anoxia may be involved in minor pathways of waterlogging tolerance. According to the IBM2 Neighbour's consensus genetic map, the major QTL on chromosome 9 is located near sucrose synthase 1, a known anaerobic response gene (McCarty et al., 1986; Springer et al., 1986; Gupta et al., 1988; Huang et al., 1994; Subbaiah and Sachs, 2003). The gene product sucrose synthase 1 was upregulated as a result of the anaerobic treatment in maize seedlings. Subbaiah and Sachs (2003) demonstrated how a simple post-translational modification of sucrose synthase by the addition/removal of phosphate can lead to potent changes in the tolerance of maize seedlings to anoxia. However, much finer mapping and a gene-specific marker are needed to prove that if this QTL actually is sucrose synthase.
Secondary QTL for waterlogging tolerance
The contributions to waterlogging tolerance of secondary QTL on chromosomes 1, 2, 3, 4, 6, 7 and 10 were all moderate with R2 values ranging from 3·9 to 14·3 % (Tables 4 and 5). Although the effects of these QTL were small, they were often detected by the CIM procedure for several traits in both environments (Fig. 1).
Eleven QTL for RL were detected in this study. Ten of the 11 QTL were detected under waterlogging-stress conditions, the exception being rl7-1 in both experiments (Fig. 1). Because RL was more sensitive to waterlogging than other traits (see Results) and its heritability was low, only one common QTL region was consistently detected under waterlogging-stress condition on chromosome 7. The results indicated that the root length QTL did not appear to contribute to root length under control conditions, and it is thus a specifically stress-responsive gene to increase root tolerance to waterlogging.
Eleven common genomic regions were associated with more than one trait in the two experimental seasons (Fig. 1, Tables 1 and 2). Seven of the eleven QTL were detected under waterlogging-stress conditions. The results indicated that the QTL region for numerous coincident traits increase plant growth under waterlogging conditions, whereas loci for only one trait may indicated a more specialized response to waterlogging.
Three of the four QTL for the WTC of SDW received positive alleles from ‘K12’, which generally has poorer phenotypic values than ‘HZ32’ (Table 1). This fact indicates that although ‘K12’ is phenotypically poor, it possesses some QTL alleles capable of increasing the trait value. Similarly, Tanksley and Nelson (1996), Bernacchi et al. (1998) and Ali et al. (2000) detected QTL alleles enhancing the trait value from a phenotypically inferior parent in tomato and rice.
Ten QTL mapped to the umc1299–umc1017 interval on chromosome 4 were detected by PH, SDW, TDW, RDW, RL and the WTC of SDW in EXP.1 and EXP.2 (Fig. 1). Out of the ten, six QTL (sdw4-2, tdw4-2, sdw4-3, ph4-2, rdw4 and tdw4-3) were found located in the same regions in both experiments. Although statistical analysis indicated that they appear to be slightly different loci, it may be possible that these QTL in fact represent the same locus because their mode of gene action was similar (Tables 4 and 5), and the peak position of a LOD score can be altered in QTL with moderate (minor) effects due to environmental interactions or statistical error. Tuberosa et al. (2002a) reported QTL for root traits in a hydroponic system using the maize cross ‘Lo964’ × ‘Lo1016’. They identified three QTL for primary root length and one for adventitious root weight located on chromosome 4. In addition, one QTL for root pulling force located on chromosome 4 was identified by Lebreton et al. (1995). The QTL on chromosome 4 for primary root length, root weight and root pulling force are adjacent to the QTL rdw4 and rl4. Mano et al. (2005) identified that the QTL controlling adventitious root formation under waterlogged conditions was also located on chromosome 4. This position overlaps with the proposed QTL for maize seedling tolerance to waterlogging in our research (Fig. 1). It appears possible that the QTL identified in our study on chromosome 4 are similar or the same as those in the region controlling these root traits. Wei and Li (2000) reported that waterlogging strongly reduced the growth of adventitious roots and dry matter accumulation of the whole root system. These results are in agreement with the root traits studied in our research (Table 1). The rl4 locus was only detected under waterlogging conditions, which may indicate that it is a specialized waterlogging-stress-responsive gene controlling root traits. However, rdw4 was detected under normal conditions, which indicated that it may be unrelated to waterlogging tolerance in this population. Moreover, Tuberosa et al. (2002b) reported that bin 1·03 harboured the major QTL for root biomass and leaf growth rate in a drain-pipe experiment carried out on the maize cross ‘DTP79’ × ‘B73’. Mano et al. (2006) also reported QTL controlling flooding tolerance in reducing soil conditions in maize seedlings using a cross of the maize inbred lines ‘F1649’ (tolerant) and ‘H84’ (sensitive). They identified a single QTL for degree of leaf injury and dry matter production located on chromosome 1 (bin 1·03–1·04). One QTL (rdw1) associated with the WTC of RDW and two QTL associated with PH were also detected on chromosome 1 (bin 1·03–1·05) in the present study (Fig. 1).The identification of similar genetic regions suggests that similarity exists among these crosses in the genetic control of root dry matter accumulation. Further research is needed to test this possibility.
Seven QTL associated with PH, RL and TDW were found within the bnlg1808 and phi034 interval on chromosome 7. Out of the seven, four QTL only associated with RL were identified under waterlogging-stress conditions. Although statistical analysis suggested that these QTL appear to be slightly separated at different loci, it may be possible that they actually represent the same locus, as also their mode of gene action was very similar. A similar result was also found on chromosome 10 (Fig. 1, Tables 4 and 5). Several genomic regions were found where the different traits measured were under related genetic control. Five QTL (ph-3, rl3-2, rdw3, tdw3-3 and tdw3-4 on chromosome 3) for PH, RL, RDW and TDW were identified under waterlogging-stress condition in EXP.2, and all appeared to be located on the same region on chromosome 3. Most of these expressed considerable negative-additivity, indicating that favourable alleles originated from ‘K12’. Although no co-location of QTL was detected between EXP.1 and EXP.2, the results indicated that QTL controlling some of the traits were on the same linkage group and sometimes close together, and genetic correlations between the traits also confirm these relations (Tables 2 and 3).
Epistasis, or interlocus interaction, is a kind of gene interaction whereby one gene interferes with the phenotypic expression of another non-allelic gene. A considerable body of classical work has strongly suggested the prevalence of an epistatic effect on quantitative traits in genetic populations (Spickett and Thoday, 1966; Allard, 1988). In the present study, most of QTL were detected in both of the two experimental years and no epistasis-by-environment interaction was detected. This indicates that a highly coherent detection of QTL was achieved between the experiments conducted in the two years. In addition, no identical epistatic QTL pairs were found to be responsible for the target traits. These results suggest that different epistatic systems control the target traits under different water-supply conditions, and that epistasis explained a considerable portion of the total genotypic variances of the measured traits and their WTC (Table 6).
In this study, most of the QTL identified by the waterlogging-response traits were located in clusters on chromosomes 3, 4, 7 and 9. Fine-mapping and trait dissection using near-isogenic lines should make it possible to determine a detailed mapping position for waterlogging tolerance QTL on chromosome 9. Once confirmed, such markers – tightly linked to waterlogging tolerance, especially to the major QTL on chromosome 9 – would facilitate the development of waterlogging-tolerant elite maize varieties and positional cloning. A concern is that the overall percentage of phenotypic variation explained by these markers remains relatively low and that other genomic regions may well also play a major role. However, this research indicates that the first steps have been set.
CONCLUSIONS
To the best of our knowledge, there has been no other report on QTL analysis of maize seedling waterlogging tolerance except for Mano et al. (2006). We have identified several QTL with genome-wide significance, suggesting that our approach is useful to elucidate the genetic mechanisms underlying maize waterlogging tolerance. The use of different populations and/or evaluation methods should allow us to detect novel QTL in further studies. Although the detected regions need to be mapped more precisely, the findings and QTL found in this study may provide useful information for marker-assisted selection (MAS) and for further genetic studies on maize waterlogging tolerance.
ACKNOWLEDGEMENTS
We thank Professor Neil Forsberg for critically reviewing the manuscript and Drs Daohua He and Faqiang Feng for the statistical analyses. This research was supported by the National Science Foundation of China (30571171), International joint research project M.O.A. (2006-G3).
LITERATURE CITED
- Ali ML, Pathan MS, Zhang J, Bai G, Sarkarung S, Nguyen HT. Mapping QTLs for root traits in a recombinant inbred population from two indica ecotypes in rice. Theoretical and Applied Genetics. 2000;101:756–766. [Google Scholar]
- Allard RW. Future direction in plant population genetics, evolution and breeding. In: Brown AHD, Clegg MT, Kahler AL, Weir BS,, editors. Plant population genetics and germplasm resources. Sunderland, MA: Sinauer Associates Inc; 1988. pp. 1–19. [Google Scholar]
- Anonymous. Studies on physio-genetic mechanism of excess soil moisture tolerance in maize. 2003 National Agricultural Technology Project, Department of Agricultural Research and Education: Indian Council of Agricultural Research (India) Wisard Project Information. [Google Scholar]
- Armstrong W. Aeration in higher plants. In: Woolhouse HW, editor. Advances in botanical research. Vol. 7. New York: Academic Press; 1979. pp. 225–232. [Google Scholar]
- Armstrong W, Drew MC. Root growth and metabolism under oxygen deficiency. In: Waisel Y, Eshel A, Kafkafi U,, editors. Plant roots: the hidden half. 3rd edn. New York: Marcel Dekker; 2002. pp. 729–761. [Google Scholar]
- Baxter-Burrell A, Chang R, Springer P, Bailey-Serres J. Gene and enhancer trap transposable elements reveal oxygen deprivation-regulated genes and their complex patterns of expression in Arabidopsis. Annals of Botany. 2003;91:129–141. doi: 10.1093/aob/mcf119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi D, Beck-Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S. Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theoretical and Applied Genetics. 1998;97:381–397. [Google Scholar]
- Boru G, van Ginkel M, Kronstad WE, Boersma L. Expression and inheritance of tolerance to waterlogging stress in wheat. Euphytica. 2001;117:91–98. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, et al. Molecular strategies for improving waterlogging tolerance in plants. Journal of Experimental Botany. 2000;51:89–97. [PubMed] [Google Scholar]
- Drew MC, Jackson MB, Giffard S. Ethylene-promoted adventitious rooting and development of cortical air spaces in roots may be adaptive responses to flooding in Zea mays L. Planta. 1979;147:83–88. doi: 10.1007/BF00384595. [DOI] [PubMed] [Google Scholar]
- Drew MC, He C-J, Morgan Page W. Programmed cell death and aerenchyma formation in roots. Trends in Plant Science. 2000;5:123–127. doi: 10.1016/s1360-1385(00)01570-3. [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene responsive-like factors regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambrell RP, Patrick WH. Chemical and microbiological properties of anaerobic soils and sediments. In: Hook DD, Crawford RMM,, editors. Plant life in anaerobic environments. Ann Arbor, MI: Ann Arbor Science; 1978. pp. 375–423. [Google Scholar]
- Ghassemi F, Jakeman AJ, Nix HA. Salinisation of land and water resources, human causes, extent, management and case studies. Sydney, Australia: University of New South Wales Press; 1995. [Google Scholar]
- Greenway H, Armstrong W, Colmer TD. Conditions leading to high CO2 (>5 kPa) in waterlogged–flooded soils and possible effects on root growth and metabolism. Annals of Botany. 2006;98:9–32. doi: 10.1093/aob/mcl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Chourey PS, Burr B, Still PE. cDNAs of two nonallelic sucrose synthase genes in maize: cloning, expression, characterization and molecular mapping of the sucrose synthase-2 gene. Plant Molecular Biology. 1988;10:215–224. doi: 10.1007/BF00027398. [DOI] [PubMed] [Google Scholar]
- He C-J, Morgan PW, Drew MC. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiology. 1996;112:463–472. doi: 10.1104/pp.112.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X-F, Binh N-Q, Prem SC, Serge Y. Complete nucleotide sequence of the maize (Zea mays L.) sucrose synthase 2 cDNA. Plant Physiology. 1994;104:293–294. doi: 10.1104/pp.104.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin SHFW, Armstrong W. Evidence for the involvement of ethylene in aerenchyma formation in adventitious roots of rice. New Phytologist. 1991;118:49–62. [Google Scholar]
- Kennedy RA, Rumpho ME, Fox TC. Anaerobic metabolism in plants. Plant Physiology. 1992;100:1–6. doi: 10.1104/pp.100.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk EJ, Wilson IW, Wilson D. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of the map from the recombination values. Ann. Eugen. 1944;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daley MJ, Lincoln SE, Etoh T. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lebreton C, Lazic-Jancic V, Steed A, Pekic S, Quarrie SA. Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. Journal of Experimental Botany. 1995;46:853–865. [Google Scholar]
- Liao C-T, Lin C-H. Physiological adaptation of crop plants to flooding stress. Proceedings of the National Science Council ROC (B) 2001;25:148–157.. [PubMed] [Google Scholar]
- Lincoln SE, Daly MJ, Lander ES. Constructing genetic linkage maps with MAPMAKER/EXP Version 3·0: a tutorial and reference manual. 3rd edn. Cambridge, MA: Whitehead Institute for Biomedical Research Technical Report; 1993. pp. 1–49. [Google Scholar]
- Liu RH, Meng JL. Map draw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas (Beijing) 2003;25:317–321. [PubMed] [Google Scholar]
- Malik AI, Colmer TD, Lambers H, Setter TL, Schortemeyer M. Short-term waterlogging has long-term effects on the growth and physiology of wheat. New Phytologist. 2002;153:225–236. [Google Scholar]
- Mano Y, Muraki M, Komatsu T, Fujimori M, Akiyama F, Takamizo T. Varietal difference in pre-germination flooding tolerance and waterlogging tolerance at the seedling stage in maize inbred lines. Japanese Journal of Crop Science. 2002;71:361–367. [Google Scholar]
- Mano Y, Muraki M, Fujimori M, Takamizo T, Kindiger B. Identification of QTL controlling adventitious root formation during flooding conditions in teosinte (Zea mays ssp. Huehuetenangensis) seedlings. Euphytica. 2005;142:33–42. [Google Scholar]
- Mano Y, Muraki M, Fujimori M, Takamizo T. Identification of QTL controlling flooding tolerance in reducing soil conditions in maize (Zea mays L.) seedlings. Plant Production Science. 2006;9:176–181. [Google Scholar]
- Materechera SA, Alston AM, Kirky JM, Dexter AR. Influence of root diameter on the penetration of seminal roots into a compacted subsoil. Plant and Soil. 1992;144:297–303. [Google Scholar]
- Mather K, Jinks JL. Biometrical genetics. 3 edn. London: Chapman & Hall; 1982. [Google Scholar]
- McCarty DR, Shaw JR, Hannah LC. The cloning, genetic mapping, and expression of the constitutive sucrose synthase locus of maize. Proceedings of the National Academy of Sciences of the USA. 1986;83:9099–9103. doi: 10.1073/pnas.83.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiology. 2005;139:949–959. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Albrecht G. Tolerance of crop plants to oxygen deficiency stress: fermentative activity and photosynthetic capacity of entire seedlings under hypoxia and anoxia. Physiologia Plantarum. 2003;117:508–520. doi: 10.1034/j.1399-3054.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Boamfa EI, Laarhoven LJJ, Harren FJM, Pors Y, Grimm B. Organ specific analysis of the anaerobic primary metabolism in rice and wheat seedlings II: Light exposure reduces needs for fermentation and extends survival during anaerobiosis. Planta. 2006;225:139–152. doi: 10.1007/s00425-006-0336-7. [DOI] [PubMed] [Google Scholar]
- Peng HP, Chan CS, Shih MC, Yang SR. Signaling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiology. 2001;126:742–749. doi: 10.1104/pp.126.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore TR Warsi. Laos Banos, Phillipines: 1998. Feb 23–27, Production of maize under excess soil moisture (waterlogging) conditions; p. 23. 2nd Asian Regional Maize Workshop PACARD (http://www.cropscience.org.au/icsc2004/poster/1/1/437_zaidi.htm?print=1. ) [Google Scholar]
- Rodrigo GS, Fernando HA, Elsa LC, Julio CC. Quantitative trait loci for grain moisture at harvest and field grain drying rate in maize (Zea mays, L.) Theoretical and Applied Genetics. 2006;112:462–471. doi: 10.1007/s00122-005-0146-5. [DOI] [PubMed] [Google Scholar]
- Saab I, Sachs MM. A flooding-induced xyloglucan endo-transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiology. 1996;112:385–391. doi: 10.1104/pp.112.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM. Molecular genetic basis of metabolic adaptation to anoxia in maize and its possible utility for improving tolerance of crops to soil waterlogging. In: Jackson MB, Black CR, editors. Interacting stresses on plants in a changing environment. Berlin: Springer-Verlag; 1993. pp. 375–393. NATO ASI Series, 16. [Google Scholar]
- Sachs MM. Gene expression in maize during anoxia. In: Basra AS, editor. Stress-induced gene expression in plants. Chur, Switzerland: Harwood Academic Publishers; 1994. pp. 87–102. [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN. Anaerobic gene expression and flooding tolerance in maize. Journal of Experimental Botany. 1996;47:1–15. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter TL, Ellis M, Lourance EV, Ella ES, Mishra Senadhira SB, Sarkarung S, Datta S. Physiology and genetics of submergence tolerance of rice. Annals of Botany. 1997;79:67–77. [Google Scholar]
- Spickett SG, Thoday JM. Regular response to selection. 3. Interaction between located polygenes. Genetic Research. 1966;7:96–121.. doi: 10.1017/s0016672300009502. [DOI] [PubMed] [Google Scholar]
- Springer B, Werr W, Starlinger P, Bennett DC, Zokolica M, Freeling M. The shrunken gene on chromosome 9 of Zea mays L. is expressed in various plant tissues and encodes an anaerobic protein. Molecular General Genetics. 1986;205:461–468. doi: 10.1007/BF00338083. [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM. Molecular and cellular adaptations of maize to flooding stress. Annals of Botany. 2003;91:119–127. doi: 10.1093/aob/mcf210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zhang Z, Zou X, Zheng Y. Functional genomics of maize submergence tolerance and cloning of the related gene Sicyp51. Science in China Series C, Life Sciences. 2005;48:337–345. doi: 10.1360/062004-27. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Nelson JC. Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into the elite breeding lines. Theoretical and Applied Genetics. 1996;92:191–203. doi: 10.1007/BF00223376. [DOI] [PubMed] [Google Scholar]
- Toojinda T, Siangliw M, Tragoonrung S, Vanavichit A. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Annals of Botany. 2003;91:243–253. doi: 10.1093/aob/mcf072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa R, Sanguineti MC, Landi P, Giuliani MM, Salvi S, Conti S. Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Molecular Biology. 2002a;48:697–712. doi: 10.1023/a:1014897607670. [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S, Sanguineti MC, Landi P, Maccaferri M, Conti S. Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Annals of Botany. 2002b;89:941–963. doi: 10.1093/aob/mcf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanToai TT, St Martin SK, Chase K, Boru G, Schnipke V, Schmitthenner AF, Lark KG. Identification of a QTL associated with tolerance of soybean to soil waterlogging. Crop Science. 2001;41:1247–1252. [Google Scholar]
- Vartapetian BB, Jackson MB. Plant adaptations to anaerobic stress. Annals of Botany. 1997;79:3–20. [Google Scholar]
- Wang DL, Zhu J, Li ZK, Paterson A. Mapping QTLs with epistatic and QTL × environmental interactions by mixed linear model approaches. Theoretical and Applied Genetics. 1999;99:1255–1264. [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer, WinQTLCart V2·0. Program in Statistical Genetics. North Carolina State University; 2002. [Google Scholar]
- Wei H-P, Li R-Q. Effect of flooding on morphology, structure and ATPase activity in adventitious root apical cells of maize seedlings. Acta Phytoecologica Sinica. 2000;24:293–297. [Google Scholar]
- Wittenburg H, Lammert F, Wang DQH, Churchill GA, Li R, Bouchard G, Carey MC, Paigen B. Interacting QTLs for cholesterol gallstones and gallbladder mucin in AKR and SWR strains of mice. Physiological Genomics. 2002;8:67–77. doi: 10.1152/physiolgenomics.00097.2001. [DOI] [PubMed] [Google Scholar]
- Xu K, Mackill DJ. A major locus for submergence tolerance mapped on rice chromosome 9. Molecular Breeding. 1996;2:219–224. [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence to tolerance to rice. Nature Letters. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Zaidi PH, Srinivasan G, Singh NN, et al. Increasing crop-water productivity through genetic improvement for tolerance to water stresses in maize (Zea mays L.). In: Fischer T, editor. New directions for a diverse planet; Proceedings for the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004; Gosford, NSW, Australia: The Regional Institute Ltd; 2004. Published online at www.cropscience.org.au . [Google Scholar]
- Zhang Z, Jiang H, Wei Z, Zheng Y. Study on enzymology in root of maize inbred after waterlogging stress. Hubei Agricultural Sciences. 2003;3:25–27. [Google Scholar]