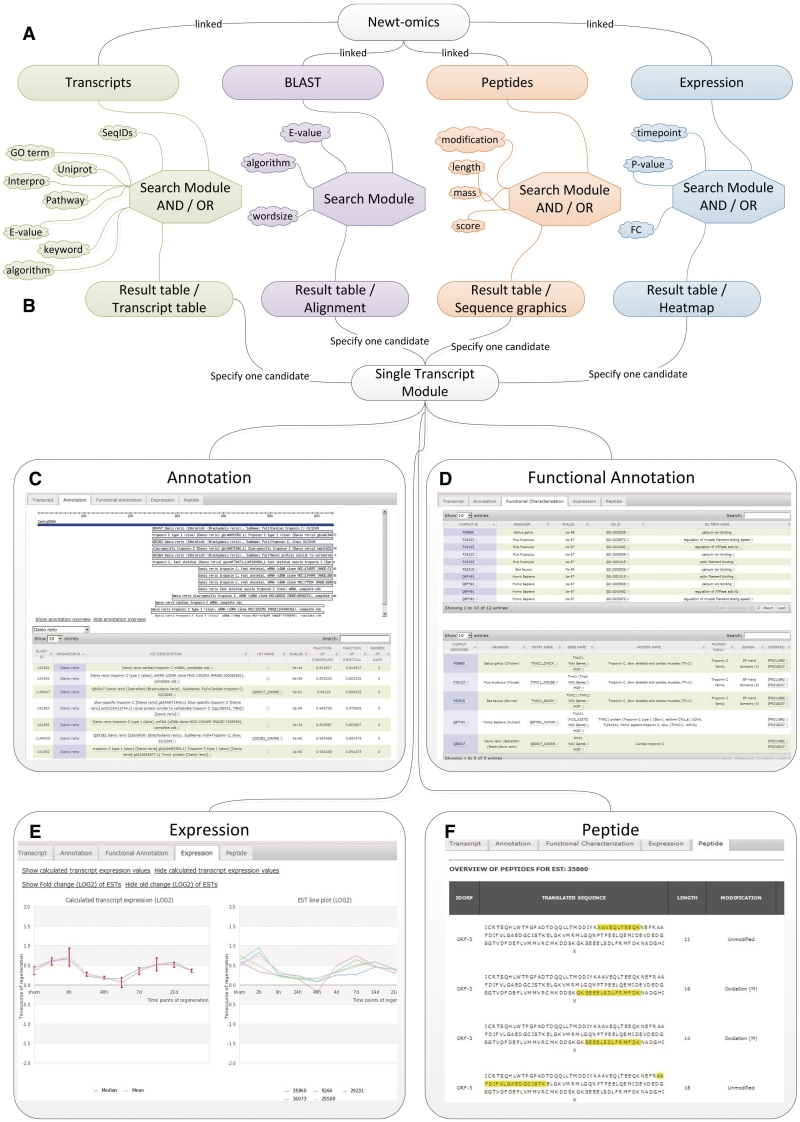
Figure 1.
The hierarchical structure of the Newt-omics Graphical User Interface (GUI) (A) The four main windows Transcripts BLAST, Peptides, Expression allow searches for single transcripts or a group of transcripts from different affiliated data sources. (B) The results tables list transcripts that link to the ‘Single Transcript Module’. All information linked to a selected transcript can be accessed via the four tabs Annotation (C), Functional Annotation (D), Expression (E) and Peptide (F).