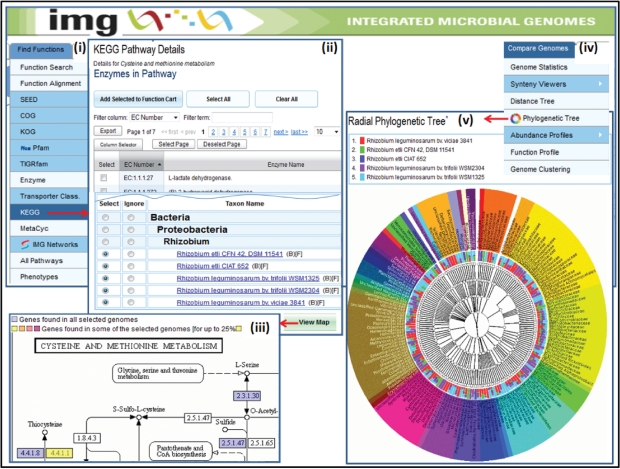
Figure 4.
Comparative analysis tools. (i) A pathway is selected from the list of KEGG pathways via the KEGG option of the ‘Find Functions’ menu, and subsequently (ii) the ‘KEGG Pathway Details’ lists its associated enzymes and the list of genomes organized phylogenetically. (iii) Once genomes are selected for comparison, the result is displayed in the context of the KEGG pathway map, with each enzyme number on the map coloured depending on the percentage of genomes with a gene associated with that enzyme. (iv) The ‘Radial Phylogenetic Tree’ is one of several tools provided for comparing genomes, and (v) allows comparing the BLAST hits of the genes of up to five user selected genomes to the genes of all the genomes in the database using a colour-coded hierarchical circular tree viewer.