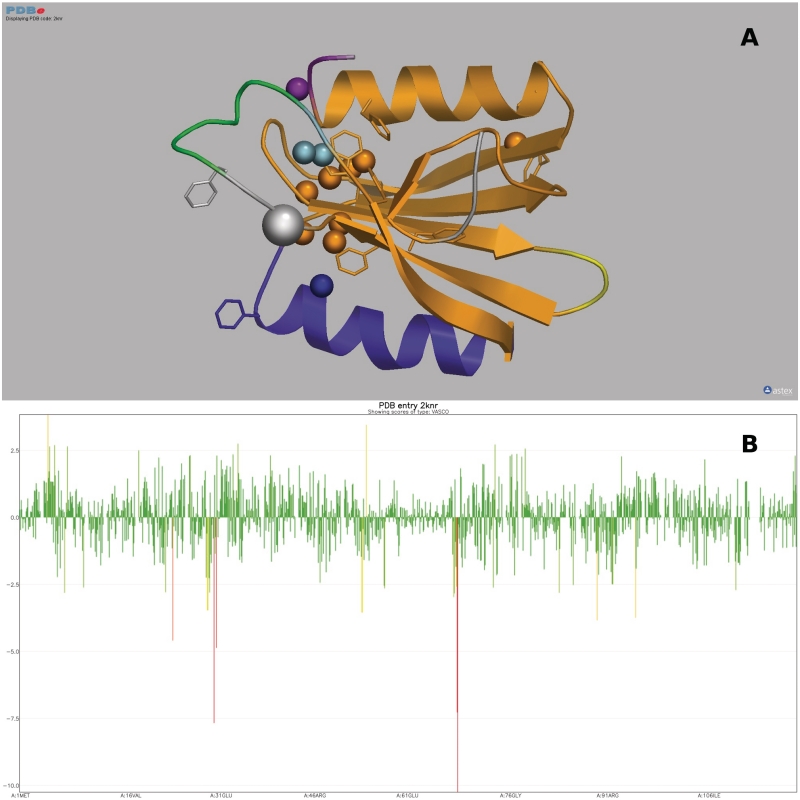
Figure 5.
Use of Vivaldi to display and analyse PDB data about NMR entries [in this example, entry 2KNR which has been discussed by Lemak et al. (33): pdbe.org/vivaldi/2knr]. (A) Results of OLDERADO (22) and VASCO (23) analysis of the 2KNR ensemble presented in a 3D structure display. OLDERADO clusters the members of an ensemble of NMR models. The representative model of the largest cluster is shown as a cartoon. The colouring of the models signifies the various structural domains indentified by OLDERADO. This analysis indicates that while the central β-sheet and the N-terminal α-helix (all in orange) are rigid with respect to each other, the second α-helix (blue) forms a separate rigid body or ‘domain’. Atoms with unusual chemical shifts as identified by VASCO are shown as van der Waals spheres. In addition, aromatic residues are shown as sticks. This structure exhibits a number of chemical shift outliers that cluster together. However, many of these outliers are near aromatic rings, which are known to strongly affect the chemical shifts values of neighbouring atoms due to ring currents. (B) 1D graph of the deviation of chemical shift values from statistical averages as judged by VASCO as a function of residue number. The height and colour of the bars convey the extent of the deviation. This graph shows that most deviations are within usual bounds (green), but some of the outliers (yellow and red) are severe (up to 10 SD) and require a plausible explanation, such as the presence of a nearby aromatic ring.