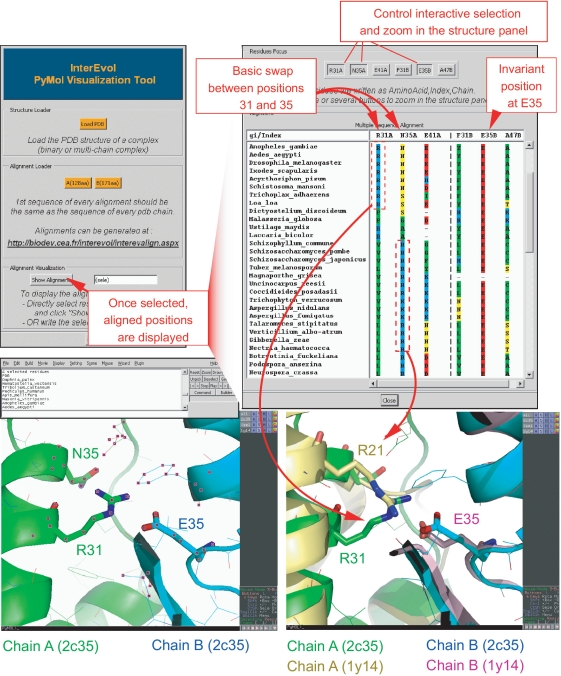
Figure 4.
Screen capture to illustrate the interest of the InterEvol PyMol plugin for diving into the structures of complex interfaces interactively with their evolutionary properties. The example focuses on the Rpb4-Rpb7 complex crystallized in both H. sapiens (pdb code:2c35) and S. cerevisiae (pdb code:1y14). The interface coordinates files were downloaded from InterEvol database in which the chains have been renumbered to match the positions in the alignment [R31, N35 and E41 in Rpb4 (2c35) stand for R44, N48 and E54 in the original PDB]. After selecting a small set of positions neighbouring at the interface it is possible to display a restricted view of the multiple sequence alignment for the 2c35 complex. Specific position can then be selected from the alignment panel and focused on in the structural panel allowing a cross talk between both dimensions. A compensatory switch between two positions of Rpb4 to maintain a salt–bridge interaction with an invariant acidic residue of Rpb7 is presented. The pre-computed alignment reveals this coordinated switch between positions 31 and 35. Retrieving the superimposed structures of the 1y14 complex reveals how structural micro-environments can adapt to allow for the plasticity of the interface in response to potentially deleterious sequence changes.