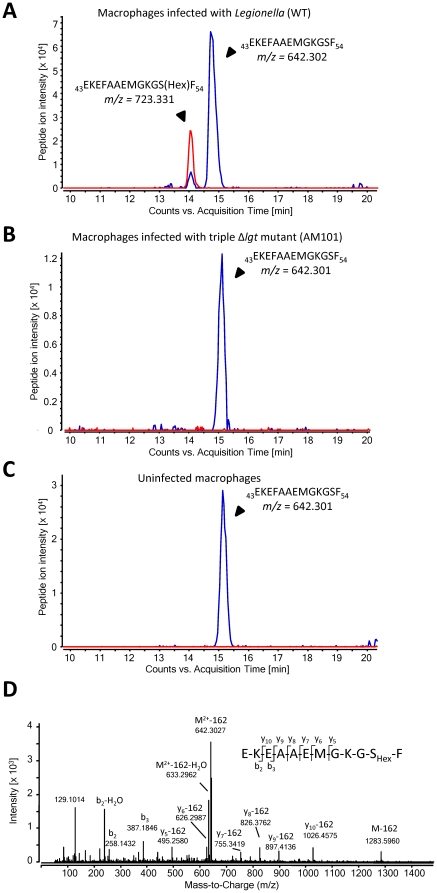
Figure 1. eEF1A is glucosylated during L. pneumophila infection at Ser53.
Mammalian eEF1A was isolated by eEF1Bα-affinity chromatography from RAW 264.7 macrophages infected with L. pneumophila wild type (A), L. pneumophila triple Δlgt-mutant AM101 (B) and uninfected cells (C). LC-MS/MS (Q-TOF) analysis revealed the extracted ion chromatograms shown. The peak at m/z = 642.3 (2+) belongs to the chymotryptic peptide 43-EKEFAAEMGKGSF-54 of eEF1A without modification. This was identified by MS/MS with a Mascot peptide Score of 72. The peptide from Legionella infected cells is partially shifted to m/z = 723.3 (2+) indicating modification by hexose (162.053 Da). The chromatograms were scanned for peptides with m/z = 642.302±10 ppm (2+) (shown in blue) and peptides with m/z = 723.331±10 ppm (2+) (shown in red). The hexose-modified form of the peptide EKEFAAEMGKGSF was exclusively detected in Legionella infected cells. Partial neutral loss of dehydrohexose (162.053 Da), typical for hexose-modified peptides, is observed already without additional collision energy in the MS scan (see Figure 1A, blue peak at 14 minutes). (D) Collision-induced dissociation MS/MS spectrum of the glucosylated peptide EKEFAAEMGKGSF (precursor m/z = 723.3 (2+); peak intensity = 2.5×104; retention time 14.06 min) identified Ser-53 as the acceptor amino acid for glucosylation (Mascot peptide Score of 60).