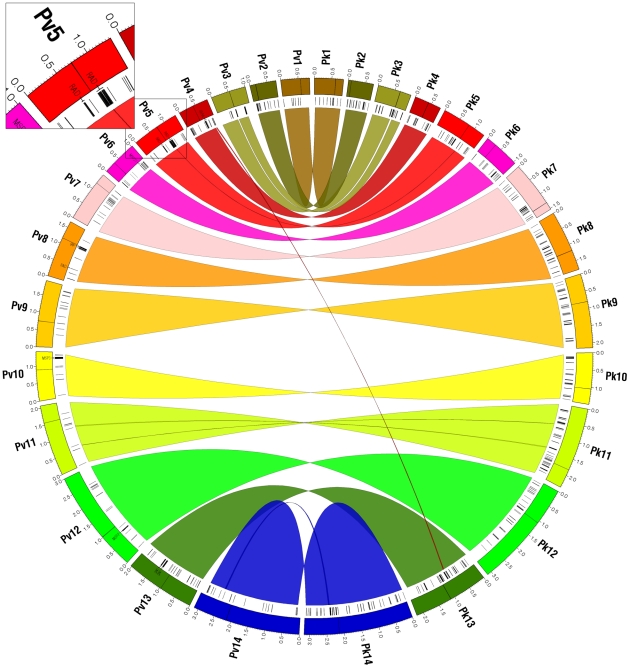
Figure 5. P. vivax and P. knowlesi share almost perfect 1-to-1 chromosomal synteny but also harbor hundreds of putative parasite-specific genes in chromosome-internal regions.
Outer segments depict the 14 nuclear chromosomes of P. vivax (left semicircle, counter-clockwise) and P. knowlesi (right semicircle, clockwise). Ribbons represent the 20 identified imperfect synteny blocks (both nested and non-nested) colored according to connected P. vivax chromosomes. Black tick marks underneath chromosomes indicate putative P. vivax-specific genes (281) and P. knowlesi-specific genes (364) located at SGRs and SBRs. Parasite-specific genes in subtelomeric regions (STRs) not shown. Text labels within chromosomes indicate parasite-specific genes mentioned in the text. The inset shows the largest identified SGR in P. vivax containing 26 RAD genes. Black lines within chromosomes indicate putative centromeres. Excluding subtelomeres, imperfect synteny blocks span complete chromosomes with only two exceptions (P. vivax chromosomes 3 and 4), but also contain many putative parasite-specific genes, particularly in P. knowlesi. Image created with Circos [56].