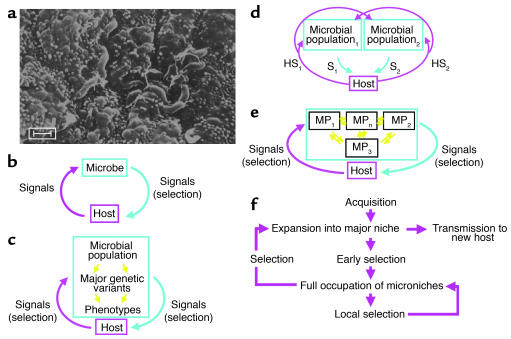
Figure 1.
Models of the cross-signaling between obligate parasites and their hosts. (a) H. pylori in the gastric (antral) niche, inhabiting the luminal mucus layer overlaying the epithelium. Scale bar: 1 μm. (b) Model 1: A coevolved microbe sends chemical and physical (contact) signals to its host. The host’s signals, including motion, temperature, and chemicals (including host-defense molecules), affect microbial growth. In a local niche, equilibrium can be reached if there is negative feedback in the cross-signaling 1, S1). The microbial signals may alter selection of growing host-cell populations (e.g., by antiapoptotic mechanisms). (c) Model 2: The microbial population includes genetic variants. Now, the host signals select for different bacterial phenotypes, and thus genotypes. (d) Model 3: Due to ongoing variation and selection, each microbe becomes a microbial population of related variants. The signals of each variant affect not only the local host signals, but also those affecting other microbial populations. This is represented by two distinct microbial populations, with each signaling the host (S1, S2) and inducing specific host signals (HS1, HS2). Increasing numbers of populations markedly and nonlinearly increase the complexity. (e) Model 4: The microbial populations are not clonal but can exchange genetic information; host selection, based on microbial genes or gene fragments rather than cells, determines the (dynamic) equilibria between the microbial populations (MPn). For the naturally competent, extensively recombining H. pylori cells, model 4 best reflects the fluidity of population structures during persistent colonization (11, S2, S3). (f) Schematic of adaptation to individual hosts. After H. pylori acquisition and expansion into a major niche, early selection allows occupation of microniches, where further local selection determines predominant populations. Both local selection and global selection determine overall population structure, and the probability of transfer of particular genotypes to new hosts. Local resource differences and barriers to diffusion of bacterial cells allow establishment of distinct subpopulations. External fluctuations, such as incomplete antibiotic treatment, can markedly change genotype distribution.