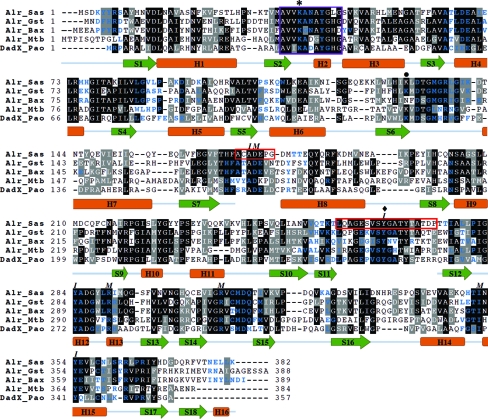
Figure 2.
Structure-based sequence alignment of alanine racemases from S. aureus (Alr_Sas), G. stearothermophilus (Alr_Gst), B. anthracis (Alr_Bax), M. tuberculosis (Alr_Mtb) and P. aeuruginosa (DadX_Pao). Identical residues are shaded black, while grey shading indicates amino acids with conserved physicochemical properties. The purple box encloses the conserved PLP-binding motif and the red boxes correspond to missing density in the AlrSas structure. I and M represent residues that form the inner and middle layers of the active-site entryway. The asterisk marks the highly conserved PLP-bound lysine, the diamond marks the location of the catalytic tyrosine and the bullet point indicates the location of a residue which is often carbamylated in alanine racemases which have a lysine at this position. The secondary structure corresponding to the amino-acid sequence of AlrSas is shown, with α-helices coloured orange and β-strands coloured green. Residues involved in the dimer interface are shown as blue letters.