Abstract
The success of genetic analyses identifying multiple loci associated with IBD susceptibility has resulted in the identification of several risk genes linked to a common cellular process called autophagy. Autophagy is a process involving the encapsulation of cytosolic cellular components in double membraned vesicles, their subsequent lysosomal degradation, and recycling of the degraded components for use by the cell. It plays an important part in the innate immune response to a variety of intracellular pathogens, and it is this component of autophagy that appears to be defective in IBD. This has lead to the hypothesis that CD may result from an impaired anti-bacterial response, which leads to ineffective control of bacterial infection, dysbiosis of the intestinal microbiota, and chronic inflammation. Several recurrent themes have surfaced from studies examining the function of autophagy-related genes in the context of IBD - with cellular context, disease status, risk variant effect and risk gene interplay all affecting the interpretation of these studies. The identification of autophagy as a major risk pathway in IBD is a significant step forward and may lead to pathway-focused therapy in the future, however there is more to understand in order to unravel the complexity of this disease.
Keywords: autophagy, inflammatory bowel disease, Crohn's disease, anti-bacterial response, mucosal immunology
Introduction
Inflammatory bowel disease (IBD) results from a complex etiology where environmental factors, genetics and intestinal microbiota combine to initiate and perpetuate chronic inflammation in the gastrointestinal tract. The two major forms of IBD are ulcerative colitis (UC) and Crohn's disease (CD), which have similar symptoms, but distinct pathological features. Recent advances in genetic analysis of IBD have resulted in significant insights into the genetic susceptibilities and underlying mechanisms of pathogenesis. To date, meta-analyses of genome wide association scan data have identified 99 risk loci - 71 CD-associated loci and 47 associated with UC, with 28 loci shared between the two types[1, 2]. It is becoming increasingly clear that these genetic risk factors are components of interacting pathways, as many of the genes encoded by these risk loci mediate common cellular functions, such as microbial recognition, autophagy, cytokine signaling, epithelial barrier maintenance, lymphocyte activation, metabolism and endoplasmic reticulum stress responses (recently reviewed in[3]). This review will focus on the recent findings linking the genes involved in the cellular process of autophagy to IBD.
Autophagy Overview
Autophagy is a conserved and highly ordered process which results in the encapsulation of cytosolic components in double membraned vesicles, their subsequent lysosomal degradation, and recycling of the degraded components for use by the cell[4]. Many different types of autophagy exist - macroautophagy, microautophagy and chaperone mediated autophagy, but for the purposes of this review we will focus on macroautophagy, hereafter termed autophagy. This process has evolved as a physiological response to cellular stress to provide nutrients in times of cellular starvation or to remove damaged organelles, protein aggregates and intracellular microbes. As a consequence, autophagy acts to ensure cellular homeostasis as a cell survival response, or if the cellular stress is severe, triggers cell death. Recent evidence has shown that autophagy also plays an important function in the innate immune response to a variety of intracellular pathogens, and it is this component of autophagy that has been proposed to be impaired in CD[5–7].
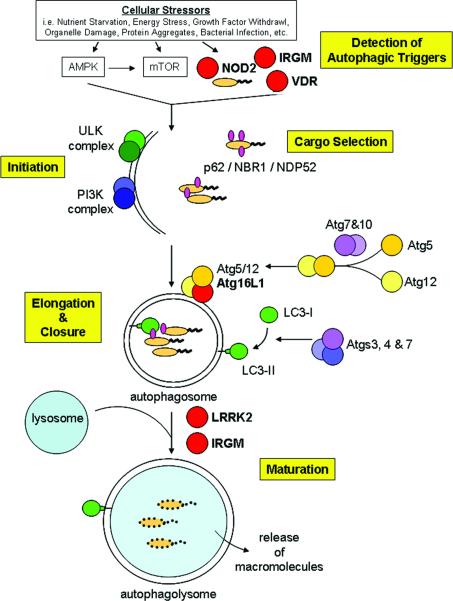
The autophagic process can be broken down into several discrete steps mediated by a cascade of kinase activation and ubiquitin-like machinery. Most of the specific components of autophagy were initially identified and characterized through studies in yeast model systems, leading to the discovery of over 32 autophagy-related (ATG) genes. Many, but not all, of the mammalian ATG homologues have been identified and the mammalian nomenclature is used in this review. This highly ordered and conserved process is can be broken down into four steps, initiation, elongation, cargo selection, and maturation (Figure 1). The molecular characterization of this process has been clearly detailed in recent reviews[4, 8, 9] and key players in the majors steps of autophagy outlined below.
Figure 1. Steps and proteins involved in autophagy.
Proteins encoded by IBD risk genes are indicated in red.
Initiation
Diverse autophagic stimuli initiate the formation of an isolation membrane through the activation of the ULK (uncorordinated-51-like kinases) kinase complex. Upon activation, the ULK complex recruits other autophagy proteins to the membrane assembly site, which include the class III phosphatidylinositol 3-kinase (PI3K) complex comprised of Beclin-1, Vps34 (vacuolar protein sorting 34), the serine/threonine kinase p150, and Barkor/mAtg14. The action of the PI3K complex generates phosphoinositides to recruit other proteins which are thought to transport membrane to the growing phagophore.
Elongation
Two ubiquitin-like conjugation systems are required for elongation of the isolation membrane. The first system is mediated by Atg7 and Atg10 which covalently link Atg12 to Atg5. This Atg12-5 conjugate interacts with Atg16L1 and tetramerizes to localize to the phagophore. The second conjugation system, comprised of Atg4, Atg7 and Atg3, modifies the cytosolic LC3 (microtubule-associated protein light chain 3) protein with the attachment of a phosphoethanolamine lipid anchor and subsequent insertion into the autophagosomal membrane. It has been proposed that the Atg12-5/16L1 complex not only promotes LC3 lipidation but also determines the site of insertion of the modified LC3 (LC3-II). The insertion of LC3 promotes cargo recruitment and autophagosomal closure. Due to the decoration of autophagosomal membranes with LC3 and the ability to detect the modified LC3-II by immunoblot, LC3 is the most commonly used marker of autophagy. It should be noted, that for simplicity we refer to LC3 modification, in mammals there are four LC3 genes and two related proteins of the GABARAP family (γ-aminobutyric acid receptor-associated proteins) that associate with autophagosomes, while in yeast there is just one gene (Atg8). The specific roles of these different mammalian Atg8 homologues are still under investigation.
Cargo Selection
The recognition and targeting of damaged organelles, protein aggregates and intracellular pathogens to autophagosomes is mediated by specific adaptor proteins interacting with LC3. Two major autophagosome adaptors are p62/sequestosome 1 (SQSTM1) and NBR1 (neighbor of BRCA1 gene 1), which directly bind ubiquitinated substrates and target them to the autophagosome through binding LC3. Another adaptor is NDP52 (nuclear dot protein 52) which may perform a more specific role in the capture of intracellular pathogens decorated with ubiquitin. The enzymes involved in the ubiquitination of autophagosome cargos are currently being investigated.
Autophagosome maturation
When autophagosome formation is complete, it matures through fusion with lysosomes to cause degradation of its contents. The fusion with lysosomes is mediated by the concerted actions of the small GTPase Rab7 and lysosome-associated membrane proteins, LAMP-1 & -2. The delivery of lysosomal hydrolases, such as the cathespsins B, D, and L as well as lysozyme, cause the degradation of the autophagosome cargo into small molecules. These small molecules are then transported into the cytosol for use by the cell or presented as antigens in the context of major histocompatability complexes to shape an adaptive immune response to infection.
Convergence of genetic and functional links between autophagy & IBD
Multiple genes that have either genetic or functional links to autophagy have IBD-associated genetic variants. These molecules include proteins involved in the detection of autophagic triggers (IRGM[10–12], NOD2[5–7], VDR[13], DAP[1, 14, 15]), orchestrating autophagosome formation (ULK1[16], ATG16L1[17, 18]) or in autophagosomal maturation (LRRK2[19]). There appears to be a stratification of these genes with the forms of IBD, with a majority of these genes thought to be specific CD susceptibility factors (ATG16L1, IRGM, NOD2, and LRRK2[3]). These findings may help shape future studies to examine additional risk factors that not only drive IBD development, but also shape the manifestation of CD vs. UC.
Interestingly, these CD-associated risk genes have an additional common functional link, as they are associated with autophagy triggered as an anti-bacterial response. This observation has lead to the hypothesis that CD may be a disease resulting from an impaired anti-bacterial response, which leads to ineffective control of bacterial infection, dysbiosis of the intestinal microbiota, and chronic inflammation. In this review, we will outline the information known about these autophagy risk genes in the context of IBD, with the focus on the function of these genes as an innate immune response to bacteria.
ATG16L1
In 2007, genome wide association scans (GWAS) identified a sole nonsynonymous single nucleotide polymorphism (SNP) in the coding region of the ATG16L1 gene associated with an increased risk of developing CD (rs2241880)[18, 20, 21]. Disease risk increases two-fold in individuals homozygous for the risk allele and linkage of this variant to IBD is stronger for CD patients with ileal disease. These findings were rapidly replicated by other groups and its association primarily with CD confirmed in recent meta-analyses[1, 2]. The prevalence of this SNP is relatively common, with as many as 45–50% of healthy individuals carrying the risk allele, emphasizing the multi-factorial nature of IBD where genetics, environment and microbiome all contribute to disease development.
Atg16L1 is an essential component of the autophagosome machinery and plays a key role for proper subcellular localization of the autophagic machinery responsible for autophagosome formation[22]. Atg16L1 undergoes self-multimerization through interactions of a central coiled coil domain forming a 800kDa complex with other autophagic molecules such as Atg5 and Atg12. A fraction of this complex localizes to the membrane but most of it remains diffuse in the cytosol. Upon induction of autophagy, the complex relocalizes to the membrane and acts as a scaffold to recruit LC3 to the membrane for lipidation. Knockdown of Atg16L1 expression in cell lines markedly decreased the targeting of intracellular bacteria to the autophagic vacuoles, demonstrating its importance in anti-bacterial autophagy[18]. Unassembled Atg16L1 gets degraded, which maintains a critical level of Atg16L1 protein in the cells for efficient autophagy. Excessive Atg16L1 inhibits autophagy, presumably through disrupting the subcellular localization of Atg16L1 and causing inappropriate localization of forming autophagosomes.
The disease-associated ATG16L1 variant results in a threonine to alanine amino acid change at position 300 (T300A) within the evolutionarily conserved WD-repeat region. In detailed studies with healthy individuals and CD patients, it was observed that Atg16L1 expression pattern at both mRNA and protein levels were not affected either by ATG16L1 genotype or inflammation state of the intestine[21]. However, the tight conservation of WD repeats among mammals led to the speculation that this amino acid substitution may alter interactions with a protein binding partner crucial for autophagy. Studies performed using truncation mutants of Atg16L1 in fibroblasts confirmed that this region is dispensable for starvation-induced autophagy[22]. Other studies have uncovered a marked impairment of antibacterial autophagy mediated by the Atg16L1 T300A variant in a cell type dependent manner[6, 23, 24]. These findings are provocative, as multiple lines of investigation support a major role in intestinal bacteria in the pathogenesis of IBD[25]. Although no single infectious agent has been identified as a causative agent for IBD[26], many of the bacteria that have been implicated in the pathogenesis of IBD, such as Mycobacterium avium subsp. paratuberculosis, adherent-invasive Escherichia coli, Listeria monocytogenes, and Yersinia enterocolitica, are also targets for autophagic clearance[8].
Cell type dependent effects of Atg16L1 have been observed in cell culture studies, as well as murine models with reduced or absent Atg16L1 expression. In studies of cell lines expressing Atg16L1 T300A, impaired anti-bacterial autophagy has been observed in epithelial cells, but not fibroblasts[6, 22–24]. When anti-bacterial autophagy was examined in primary human cells homozygous for the risk allele, conflicting results were observed, possibly dependent on donor disease status. In one study examining macrophages and dendritic cells from healthy individuals, no significant differences were observed between healthy and risk genotypes[6]. This contrasts with the findings of two other groups examining cells derived from CD patients, who observed significant impairment of autophagy in lymphoblasts or dendritic cells activated by muramyl dipeptide, a component of bacterial cell wall and activating ligand of NOD2 (nucleotide binding oligomerization domain 2), a bacterial sensor and CD risk gene[5, 7]. Interestingly, this impairment was not observed in dendritic cells stimulated with a different component of the bacterial cell wall[5], suggesting further complexity to the regulation of autophagy in the context of IBD.
To investigate the role of ATG16L1 in an in vivo setting, two different approaches were taken by investigators. One group generated mice with a functional knockout of the Atg16L1 protein (Atg16L1KO)[27]. Autophagy plays an essential role in the survival of newborn mice and autophagy-deficient mice die within 24 hours of birth[27–29]. Therefore, these investigators generated fetal liver chimeric mice, resulting in wildtype mice with the hematopoietic system of Atg16L1KO mice. Using these Atg16L1KO chimeric mice, the investigators were able to study the impact of Atg16L1 deficiency on immune cell function. Atg16L1KO macrophages were unable to activate autophagy, but had enhanced IL-1β and IL-18 production when stimulated with bacterial endotoxin (lipolysaccharide or lipid A) or other activators of the inflammasome. This hypersensitivity was also observed when these mice were tested in an experimental colitis model induced by dextran sulfate sodium (DSS). Colitis induction was exacerbated in Atg16L1KO chimeric mice, with increased ulceration and lymphocytic infiltration, which correlated with high serum levels of the pro-inflammatory cytokines IL-1β, IL-6 and IL-18. Hyper-production of IL-1β was also observed in peripheral blood mononuclear cells from CD patients and healthy volunteers genotyped for the ATG16L1 risk variant[30]. However, the effect observed in the human cells was only induced when the cells were stimulated with the NOD2 ligand MDP, but not other bacterial stimuli, possibly reflecting a difference between absence of Atg16L1 function and altered Atg16L1 function as a result of the T300A substitution. These findings demonstrate a role for autophagy and Atg16L1 function in the regulation of pro-inflammatory cytokine production in response to bacterial stimulation.
In a second model, investigators generated mouse lines with the expression of Atg16L1 reduced to ~30% of normal levels (Atg16L1 hypomorph; Atg16L1HM)[31]. Although the overall morphology of the intestine or colon of these mice was not affected and no spontaneous inflammation was observed, closer analysis uncovered striking changes in Paneth cells of the small intestine. These changes include morphological abnormalities, functional changes and transcriptional alterations. Paneth cells are highly secretory cells at the base of the crypts of Lieberkühn which play an important role in mucosal defense through the production and secretion of anti-microbial peptides, lysozyme, and inflammatory mediators into the gut lumen. Transcriptionally, the Paneth cells of Atg16L1HM mice produced high levels of the immunoregulatory adipocytokines, leptin and adiponectin, both of which have been linked to a pathologic hallmark of CD termed “creeping fat”[32]. These cells also displayed abberant packaging and exocytosis of anti-microbial granules. Strikingly similar Paneth cell abnormalities were also observed in CD patients homozygous for the ATG16L1 risk allele, indicating that this mouse model may reflect important aspects of human disease[31].
In follow-up studies of the Atg16L1HM mice, it was discovered that the Paneth cell abnormalities were triggered by infection with a specific strain of murine norovirus (MNV CR6) which generates persistent infection[33]. When these mice were re-derived by embryo transfer into an enhanced barrier facility, Paneth cell morphology of the Atg16L1HM mice was indistinguishable from wild-type mice. Additionally, these mice had similar responses to DSS-induced intestinal injury as wild-type mice, indicating that reduction of Atg16L1 expression (or autophagy) alone is not sufficient for the pathologic changes. Further investigation discovered that prior, persistent infection with MNV of lamina propria cells induced not only changes in Paneth cell morphology and transcriptional profiles of these cells, but also resulted in an aberrant intestinal injury response with hallmarks of human CD. Rather than the typical surface ulceration of the transverse and distal colon observed in the DSS colitis model, in the virally infected Atg16L1HM mice, DSS administration also resulted in ileal pathology, with mucosal atrophy, muscular hypertrophy, increased lymphoid aggregates, development of subserosal fibrosis and hyperplasia. This aberrant injury response was found to be dependent on TNFα, IFNγ and the commensal microbiota, three factors closely associated with CD. These findings highlight the multi-factorial nature of CD development, where carriage of a genetic risk factor alone is not sufficient to cause disease.
NOD2
The first gene associated with CD susceptibility was NOD2 (nucleotide-binding, oligomerization domain 2), which encodes an intracellular bacterial sensor of the NLR (NOD-like receptor) family[34]. It was originally identified by two separate groups using distinct genetic approaches and has been extensively confirmed as a CD risk gene in individuals of European descent. The three NOD2 variants most commonly associated with CD are two missense mutations, arginine 702 to tryptophan (R702W, rs2066844) and glycine 908 to arginine (G908R, rs2066845) as well as a frameshift mutation resulting from a cytosine insertion after amino acid 1007 (L1007fs, rs41450053). These main variants result in a significant reduction or loss of NOD2 function. Heterozygous carriage of one the major risk alleles confers a 2.4-fold increased risk of CD, while homozygous or compound heterozygous individuals have a 17.1-fold increased risk in individuals of European decent. Interestingly, no association of NOD2 variants has been observed in Asian or sub-Saharan African populations. Due to the multi-factorial nature of disease onset, the carriage of NOD2 variants is not sufficient to cause CD, but they are associated with disease characterized by an earlier age of onset, ileal location and stricture formation[3].
NOD2 is an intracellular protein expressed in a variety of immune and non-immune cells where it is involved in recognition and defense against pathogens. Muramyl dipeptide (MDP) is a ubiquitous component of the bacterial peptidoglycan cell wall and the best defined activating ligand of NOD2. Upon MDP stimulation, NOD2 activates the mitogen activated protein kinase (MAPK) cascade, as well as the transcription factor NFκB, resulting in the transcription and release of pro-inflammatory mediators and anti-microbial products. NOD2 also induces nitric oxide production which has a direct microbicidal effect[35]. In mice, NOD2 deficiency results in reduced anti-microbial defensin expression, increased susceptibility to pathogens[36, 37] and alterations in the types and amount of commensal bacteria[38]. Several of these changes (reduced β-defensin expression and altered bacterial microbiome) have also been observed in individuals with NOD2-associated CD[39–41].
Recent studies by three different groups have uncovered autophagy as an additional anti-microbial response triggered by NOD2[5–7]. These studies linking NOD2 to autophagy uncovered a novel risk gene interaction between NOD2 and ATG16L1 and reinforces the concept that IBD susceptibility is due to a dysregulation of a central pathway, rather than dysfunction of one specific gene. Autophagy induction by NOD2 is dependent upon Atg16L1 expression and has been observed in multiple cell types[5–7]. In epithelial cells, NOD2 was shown to interact with Atg16L1 to guide Atg16L1 to bacterial entry sites in the plasma membrane to initiate the autophagic response[7]. Previous studies have demonstrated that the NOD2 L1007fs risk variant, unlike wild-type NOD2, does not localize to the plasma membrane[42, 43]. In cells expressing NOD2 L1007fs, formation of bacteria-targeted autophagosomes was impaired, with both NOD2 L1007fs and Atg16L1 mislocalized to the cytosol, indicating that this membrane localization is a key contributor to the rapid autophagic clearance of bacteria[7].
Other studies examining NOD2-mediated autophagy in dendritic cells demonstrated an additional role for this process in MHC class II antigen presentation to T lymphocytes[5]. Using primary immature human dendritic cells, the investigators demonstrated that MDP induced autophagosome formation, MHC class II upregulation and antigen presentation in a NOD2-dependent manner. These processes also required the expression of known autophagy proteins, such as Atg5, Atg7 and Atg16L1, and the NOD2 signaling mediator RIPK2 (receptor-interacting serine-threonine kinase 2; RIP2/RICK). Similar requirements for NOD2-mediated autophagy induction were also observed in intestinal epithelial cells and extended to demonstrate that signaling independent of IKKs (IκB kinases) and NFκB is required for NOD2-mediated autophagy induction[6]. The effect of disease-associated variants was also examined in primary dendritic cells from genotyped CD patients homozygous or compound heterozygous for the major NOD2 risk variants[5]. These cells with disease-associated NOD2 variants showed an impairment of autophagy induction, MHC class II antigen presentation and bacterial handling of Salmonella typhimurium or adherent invasive Escherichia coli infection. Interestingly, these defects appear to be specific to NOD2-mediated autophagy, as autophagy induction was not impaired when these cells were stimulated with a different bacterial ligand (Pam3CSK4) or an autophagy activating drug (rapamycin).
Another study indicates that autophagy may play a role in the activation of NOD2-dependent signal transduction pathways[6]. This is similar to the autophagic delivery of microbial ligands to other microbial sensors which reside in intracellularly, such as Toll-like receptors 7–9 found in endosomes[44, 45]. In studies of epithelial cells, it was found that MDP-stimulated activation of NFκB was significantly reduced when autophagy was blocked by chemical inhibitors or RNA interference (RNAi)-mediated knockdown of Atg16L1 expression[6]. These results suggest autophagy is an additional, novel mechanism for MDP intracellular trafficking, which may contribute to a positive feedback loop during intracellular bacterial infection. However, this effect appears cell type and cell function dependent, as MDP-stimulated NFκB activation in human epithelial cells or primary human monocytic-derived cell types (macrophages and dendritic cells) was similar to cells expressing wild-type Atg16L1, but MDP-enhanced bacterial killing in epithelial cells was dramatically impaired. Future studies carefully examining these different cell type and cell function effects may identify key locations and defects related to disease pathogenesis.
In summary, the intersection of NOD2 with autophagy presents a multi-pronged approach in the defense against intracellular bacteria by: 1) guiding ATG16L1 to the bacterial entry site to initiate autophagy, 2) through presentation of MHC class II antigens to T cells and shaping adaptive immune responses, and 3) by contributing to the activation of pro-inflammatory signaling mediated by NOD2. It should be noted, that similar to the studies of Atg16L1, the cell type and experimental context of study are important to the effects of disease-associated risk variants on autophagy and NOD2 function.
IRGM
Immunity related guanosine triphosphatases (IRGs) are a family of proteins essential for the elimination of several different intracellular pathogens in most mammals[46]. In contrast to mice which have 21 IRG family members, humans carry only two IRG genes (IRGC and IRGM), with only IRGM having a role in host defense. Human IRGM has five different splice variants (IRGMa-e); but, due to low levels of expression and differential gene structure in animal models, elucidation of the specific functions of the IRGM isoforms has been challenging and incomplete. A recent paper suggests that these isoforms may have distinct roles in promoting autophagy, mitochondrial fission, inflammation and cell death[47]. IRGM expression has been widely detected in many different human cell lines, as well as primary cells from the colon, small intestine, peripheral blood leukocytes and monocytes.
The connection between IRGM and autophagy was first established based on its role in clearance of Mycobacterium tuberculosis[47]. Functionally, IRGM has been implicated in autophagy induction, as well as autophagosomal maturation, and is associated with the autophagosome membrane[47, 48]. Reduction of IRGM expression resulted in increased survival of intracellular bacteria such as M. tuberculosis, Salmonella typhimurium and the CD-associated adherent-invasive Escherichia coli strain LF82[24, 47, 49, 50]. Likewise, modest overexpression of IRGM enhanced autophagic bacterial clearance, but high levels of expression led to intracellular bacterial accumulation due to an overload in the lysosomal compartments[49], indicating that autophagy is regulated by a critical threshold of IRGM expression.
Multiple CD-associated SNPs have been identified around the IRGM locus, with the strongest replication signal associated with two SNPs (rs13361189 and rs4958847) flanking the coding region of IRGM[11, 20]. However, these disease-associated SNPs did not result in amino acid sequence changes and led to speculation that they may affect IRGM expression, splicing or translational rate. Further analysis uncovered a 20kb deletion polymorphism immediately upstream of IRGM in perfect linkage disequilibration with a disease-associated SNP (rs1336119)[50]. Additionally, these IRGM haplotypes were found to be tagged with another exonic synonymous SNP (rs10065172; c.313C>T) in the coding region of IRGM, which facilitated analysis of the risk (IRGMT) and protective (IRGMC) variants. The variants were observed to have significantly different levels of expression which was also cell type dependent. Furthermore, another study suggests that the presence of the deletion polymorphism not only affects the overall expression of IRGM, but also may alter the relative proportion of IRGM splice variants[46].
A recent study confirmed that the expression of IRGM risk variants is altered in CD and determined that the expression of IRGM is regulated by the binding of microRNAs (miRNAs; miR-196) to the region of IRGM where the c.313C>T disease-associated SNP is found[49]. Binding studies between miR-196 and the risk (IRGMT) or protective (IRGMC) forms of IRGM indicate a decreased binding of miR-196 to IRGMT, resulting in higher expression levels of IRGMT in cell lines. In vivo findings concur with these results as increased miR-196 expression was observed in the inflamed mucosa of CD patients, correlating with specific decreases in IRGMC expression in intestinal epithelial cells, but not IRGMT expression. This sustained IRGM expression resulted in an increase in targeting of AIEC bacteria by autophagy, but defective autophagosome maturation, causing ineffective pathogen clearance. Therefore, it appears that a delicate balance of IRGM expression regulated by miRNAs is required to bring about efficient pathogen clearance and maintain a healthy gut environment.
LRRK2
Genetic variants of LRRK2 (leucine-rich repeat kinase 2) have been associated with CD, Parkinson's disease and leprosy[19, 51–53]. Alterations in autophagy have been proposed as potential pathogenic mechanisms for all three disorders. The CD-associated SNP (rs1175593) is located upstream of the coding sequence of LRRK2[19]. However, the consequence of this SNP on LRRK2 function or expression has not yet been determined. LRRK2 is primarily expressed in the kidney and circulating leukocytes, with expression enhanced by exposure to IFNγ or microbial products (LPS, lentiviral particles, ssRNA analogue R837, CpG DNA)[54, 55]. Consistent with this association, LRRK2 expression levels were found to be significantly upregulated in colonic biopsy specimens from inflamed tissues of CD patients[54]. Interestingly, in contrast to many of the other autophagy risk genes, LRRK2 expression was not observed in the intestinal epithelium, but only lamina propria leukocytes. LRRK2 colocalizes with two autophagy proteins (p62 and LC3) and has been suggested to play a crucial role in maintaining autophagic equilibrium[56].
ULK1
ULK1 is a component of an essential protein complex involved in initiation of autophagy. Recent genetic analysis of ULK1 using two different approaches in separate cohorts (SNP analysis and transmission disquilibrum testing) identified a single SNP (rs12303764) significantly associated with CD[16]. This SNP is located 43bp after the end of exon 16, but it is not known yet what effect this SNP will have on the function or expression of ULK1. It is clear, however, that alterations in ULK1 will have widespread effects on autophagy induction, as it is a central integrator of multiple autophagy-inducing stimuli. For example, AMPK (AMP-activated protein kinase) and mTOR (mammalian target of rapamycin) are two major regulators of ULK1 activity in times of nutrient starvation or energy stress[4]. ULK1 activity is regulated by a complex array of multiple phosphorylation and dephosporylation events which influence the binding of regulatory and effector autophagy proteins[57, 58]. It will be interesting to see how the carriage of this SNP will impact the therapeutic use of autophagy-targeting drugs in CD.
VDR
Linkage studies have shown that polymorphisms in VDR (vitamin D receptor) are associated with IBD susceptibility (both CD and UC) in non-European populations[13, 59–61]. Based on other studies identifying vitamin D deficiency as a risk factor for CD[62] and the increased susceptibility of VDR-deficient mice to colitis observed in multiple models[63–66], we would predict that these polymorphisms would negatively affect VDR function. Supporting data specifically connecting disease-associated VDR variants to altered autophagy induction has not been published; however, several studies link vitamin D to the induction of anti-microbial autophagy[67]. Vitamin D can regulate autophagy at multiple steps of the autophagic process, through modulation of calcium fluxes and autophagy gene expression. A recent study demonstrated that the expression of NOD2 is regulated by the activated VDR[68] and VDR binding sites have been identified in the promoter of another IBD risk gene, PTPN2 (protein tyrosine phosphatase, non-receptor 2)[69], again highlighting the functional interaction of IBD risk genes.
Application of autophagy modulation to IBD therapy
The previously described studies clearly suggest a link between alterations in autophagy with susceptibility to IBD. The question remains whether this information can be exploited therapeutically to benefit individuals with IBD. The first question is whether there are autophagy-modulating compounds available which are both effective and are safe to use in a clinical setting. Fortunately, autophagy is already a therapeutic target or proposed target for several other diseases, such as cancer, muscle atrophy, myopathy, cardiac disease, pancreatitis, liver disease, stroke, type 2 diabetes, Parkinson's disease, Lafora disease, Huntington's disease, and Alzheimer's disease[70]. Additionally, prospective chemical compound screens have identified several FDA-approved drugs which target autophagy[71, 72] and specific bioactive food components have been implicated in autophagy stimulation[73] (Table I). Many of these compounds target mTOR, a major integrator of a plethora of autophagy-inducing stimuli, including nutrient starvation, growth factor depletion and endoplasmic reticulum stress. A better understanding of what autophagy signaling mechanisms are altered in IBD will assist in the identification and rational design of “IBD effective” compounds.
Table I.
| Category / Target | Name | Disease Application |
|---|---|---|
| mTOR inhibitor / Immunosuppressant | Amiodarone | Angina, cardiac arrhythmia |
| Everolimus | Lymphoid cancers | |
| Niclosamide | Anti-helminthic | |
| Perhexiline | Angina | |
| Rapamycin | Lymphoid cancers | |
| Sirolimus | Lymphoid cancers | |
| Calcium channel blockers | Aminodarone | Hypertension, angina, cardiac arrhythmia |
| Loperamide | Diarrhea associated with gastroenteritits and IBD | |
| Niguldipine | Angina, hypertension, Raynaud's phenomenon | |
| Nicardipine | Hypertension, angina, cardiac arrhythmia | |
| Anti-psychotic | Fluspirilene | Schizophrenia |
| Pimozide | Huntington's disease, schizophrenia, chronic psychosis | |
| Trifluoperazine | Anti-psychotic, Huntingtion's disease | |
| Dietary inducer | Curcumin | Brain cancer |
| Genistein | Ovarian cancer | |
| Quercetin | Colon cancer | |
| Resveratrol | Colon, colorectal, ovarian, lung and salivary gland cancers | |
| Vitamin C | Lung cancer | |
| Vitamin D3 | Head and neck cancers |
Results of autophagy activating compounds in animal colitis models, CD case reports and clinical IBD trials are encouraging, although limited. An initial study reported that everolimus (a mTOR inhibitor) significantly reduced the severity of chronic colitis in an interleukin-10 deficient (IL-10KO) mouse model[74]. Similar findings were found in two case reports demonstrating that treatment with mTOR inhibitors (everolimus and sirolimus) resulted in marked and sustained improvement of disease in refractory CD patients[75, 76]. However, when everolimus was tested on CD patients with moderate-to-severe disease in a larger clinical trial, everolimus treatment was not significantly effective, only 22% of patients maintained steroid-free remission[77]. It should be noted, however, that the genotype of the patients enrolled in the study was unknown and is likely to be a major factor in the success of this type of treatment.
Other studies examined the effect of a dietary autophagy inducer, vitamin D, on experimental colitis and in clinical trials of CD. In the DSS colitis model, administration of vitamin D or a VDR agonist (BXL062) resulted in the amelioration of the signs and symptoms of colitis[78, 79], while mice fed a vitamin D-deficient diet were more susceptible to DSS-induced colitis[80]. Clinically, a small prospective clinical study reported significant improvements of CD Activity Index (CDAI) scores and reductions in serum inflammatory markers in CD patients treated with an active form of vitamin D[81]. However, the effect of oral vitamin D supplementation in a randomized double-blind placebo study on CD relapse rate demonstrated a reduced relapse rate (29% to 13%), but the reduction did not reach statistical significance in this modestly powered study[82]. While promising, it is unclear if the efficacy of vitamin D is due solely to its autophagy stimulatory effects or in combination with other immune modulatory functions. The use of “natural” products to enhance autophagy is attractive, as they have fewer side effects and may be more effective for long term treatment of chronic conditions such as IBD.
Future directions
It is clear that we have just begun to understand the complexity of inflammatory bowel disease susceptibility pathogenesis, but recent studies demonstrate that autophagy plays a major role in this complexity. Several recurrent themes have surfaced from studies examining the function of autophagy-related genes in the context of IBD - with cellular context, disease status, risk variant effect and risk gene interplay all affecting the interpretation of these studies. The cell type context of the disease risk gene dramatically affects the overall effect of disease-associated variants. For example, several investigators have highlighted alterations in Paneth cell function as a primary defect resulting from carriage of autophagy risk variants in ATG16L1 and NOD2, however expression of IRGM in Paneth cells is not altered by risk variants and LRRK2 does not appear to be significantly expressed in this cell type. Additionally, the effect of ATG16L1 T300A produced dramatically different effects on the autophagic killing of intracellular bacteria in epithelial cells and IL-1β production in macrophages. The elucidation of the cell type specific functions of these risk genes will provide insights into the cellular basis of disease development.
Likewise, the disease context of these risk genes also appears to affect their function, with discrepancies in the effect of ATG16L1 T300A variant on MDP-stimulated autophagy observed in dendritic cells from healthy individuals (no effect) and CD patients (autophagy blockade). Similarly, colitis studies in ATG16L1HM mice demonstrated that prior, persistent viral infection is required to produce the Paneth cell defects associated with ATG16L1 T300A in CD patients. These findings underscore that IBD is a complex disease with multiple risk factors that are not sufficient individually to result in disease. The disease-dependent phenotype of risk genes provides an opportunity to examine the interaction of environmental risk factors with IBD genetics.
Although many laboratories are actively studying IBD risk genes and the effects disease-associated variants have on their function, much more needs to be known about the specific roles these genes and the autophagic process plays normally in immune responses, shaping the microbiota, and inflammatory processes. Some risk genes do not have clearly defined functions in healthy individuals or the disease-associated polymorphism does not affect the final sequence of the protein encoded by the gene, adding significant challenges to the analysis of these risk factors. In many cases, global absence of risk gene expression does not accurately reflect the effect of disease-associated variants. Therefore, the development of genetically engineered mouse models and use of genotyped material from both healthy and IBD patients will be necessary for the analysis of disease-associated changes.
Adding to the complexity of genetic studies is accumulating evidence that risk genes affect the expression and function of other IBD risk genes. Functional cross-talk between ATG16L1 and NOD2 has been demonstrated with disease-associated variants in each gene affecting the function of the other. Similarly, the expression of NOD2 and other autophagy genes can be increased by VDR stimulation. These findings, as well as others, suggest that the functional interplay between genetic risk factors indicate that overall IBD susceptibility may be better assessed by carriage of the total number of disease-associated polymorphisms in a risk pathway, as opposed to homozygosity of an individual risk gene.
The success of genetic analysis in identifying multiple loci associated with IBD susceptibility has resulted in the identification of common disease-associated pathways suitable for therapeutic manipulation. Although promising for the advent of personalized medicine, more detail is required to determine the application of genetic risk to successful IBD therapy. Only approximately 25% of the heritable risk of IBD has been identified to date, so additional genetic studies focusing on uncommon alleles and more intricate systems analysis of disease-associated pathways to identify additional components of these risk pathways is warranted. The association of genetics with disease phenotype (location, age of onset, stricturing, etc.) will also provide insights into pathogenic mechanisms of disease. Importantly, the incorporation of genetic risk information in future clinical trials will enhance both the design and analysis of pathway targeted therapies. Although the identification of autophagy as a major risk pathway in IBD is a significant step forward, there is more to understand before completely unraveling the complexity of this disease.
Acknowledgments
Grant Support: This work was supported by a National Institutes of Health research grant, R01DK082437 to C.M. and the Howard Hughes Medical Institute “Med Into Grad” initiative.
References Cited
- 1.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42(12):1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140(6):1704–12. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16(1):90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 6.Homer CR, Richmond AL, Rebert NA, et al. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139(5):1630–41. 1641, e1–2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11(1):55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 8.Sumpter R, Jr., Levine B. Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol. 2010;21(7):699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 10.Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn's disease. Autophagy. 2007;3(6):649–51. doi: 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- 11.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39(7):830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts RL, Hollis-Moffatt JE, Gearry RB, et al. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9(6):561–5. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 13.Simmons JD, Mullighan C, Welsh KI, Jewell DP. Vitamin D receptor gene polymorphism: association with Crohn's disease susceptibility. Gut. 2000;47(2):211–4. doi: 10.1136/gut.47.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koren I, Reem E, Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol. 2010;20(12):1093–8. doi: 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Kuester D, Guenther T, Biesold S, et al. Aberrant methylation of DAPK in long-standing ulcerative colitis and ulcerative colitis-associated carcinoma. Pathol Res Pract. 2010;206(9):616–24. doi: 10.1016/j.prp.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Henckaerts L, Cleynen I, Brinar M, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn's disease. Inflamm Bowel Dis. 2010;17(6):1392–7. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 17.Cummings JR, Cooney R, Pathan S, et al. Confirmation of the role of ATG16L1 as a Crohn's disease susceptibility gene. Inflamm Bowel Dis. 2007;13(8):941–6. doi: 10.1002/ibd.20162. [DOI] [PubMed] [Google Scholar]
- 18.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Welcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 22.Fujita N, Itoh T, Omori H, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuballa P, Huett A, Rioux JD, et al. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3(10):e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapaquette P, Glasser AL, Huett A, et al. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2009;12(1):99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1720–28. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 26.Macfarlane GT, Blackett KL, Nakayama T, et al. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15(13):1528–36. doi: 10.2174/138161209788168146. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 30.Plantinga TS, Crisan TO, Oosting M, et al. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011 doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 31.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffler A, Herfarth H. Creeping fat in Crohn's disease: travelling in a creeper lane of research? Gut. 2005;54(6):742–4. doi: 10.1136/gut.2004.061531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annu Rev Med. 2007;58:401–16. doi: 10.1146/annurev.med.58.061705.145024. [DOI] [PubMed] [Google Scholar]
- 35.Totemeyer S, Sheppard M, Lloyd A, et al. IFN-gamma enhances production of nitric oxide from macrophages via a mechanism that depends on nucleotide oligomerization domain-2. J Immunol. 2006;176(8):4804–10. doi: 10.4049/jimmunol.176.8.4804. [DOI] [PubMed] [Google Scholar]
- 36.Biswas A, Liu YJ, Hao L, et al. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci U S A. 2010;107(33):14739–44. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307(5710):731–4. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 38.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106(37):15813–8. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(1):179–84. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid M, Fellermann K, Wehkamp J, et al. The role of defensins in the pathogenesis of chronic-inflammatory bowel disease. Z Gastroenterol. 2004;42(4):333–8. doi: 10.1055/s-2004-813072. [DOI] [PubMed] [Google Scholar]
- 41.Wehkamp J, Harder J, Weichenthal M, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53(11):1658–64. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnich N, Aguirre JE, Reinecker HC, et al. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-{kappa}B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–6. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lecine P, Esmiol S, Metais JY, et al. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282(20):15197–207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 44.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28(6):799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HK, Lund JM, Ramanathan B, et al. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 46.Bekpen C, Xavier RJ, Eichler EE. Human IRGM gene “to be or not to be”. Semin Immunopathol. 2009;32(4):437–44. doi: 10.1007/s00281-010-0224-x. [DOI] [PubMed] [Google Scholar]
- 47.Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 48.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302(5645):654–9. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 49.Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43(3):242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 50.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40(9):1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paisan-Ruiz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Burgunder JM, An X, et al. LRRK2 R1628P variant is a risk factor of Parkinson's disease among Han-Chinese from mainland China. Mov Disord. 2009;24(13):1902–5. doi: 10.1002/mds.22371. [DOI] [PubMed] [Google Scholar]
- 53.Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Gardet A, Benita Y, Li C, et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol. 2011;185(9):5577–85. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hakimi M, Selvanantham T, Swinton E, et al. Parkinson's disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J Neural Transm. 2011;118(5):795–808. doi: 10.1007/s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alegre-Abarrategui J, Christian H, Lufino MM, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18(21):4022–34. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JW, Park S, Takahashi Y, et al. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5(11):e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dresner-Pollak R, Ackerman Z, Eliakim R, et al. The BsmI vitamin D receptor gene polymorphism is associated with ulcerative colitis in Jewish Ashkenazi patients. Genet Test. 2004;8(4):417–20. doi: 10.1089/gte.2004.8.417. [DOI] [PubMed] [Google Scholar]
- 60.Naderi N, Farnood A, Habibi M, et al. Association of vitamin D receptor gene polymorphisms in Iranian patients with inflammatory bowel disease. J Gastroenterol Hepatol. 2008;23(12):1816–22. doi: 10.1111/j.1440-1746.2008.05525.x. [DOI] [PubMed] [Google Scholar]
- 61.Pei FH, Wang YJ, Gao SL, et al. Vitamin D receptor gene polymorphism and ulcerative colitis susceptibility in Han Chinese. J Dig Dis. 2011;12(2):90–8. doi: 10.1111/j.1751-2980.2011.00483.x. [DOI] [PubMed] [Google Scholar]
- 62.Cantorna MT. Vitamin D and its role in immunology: multiple sclerosis, and inflammatory bowel disease. Prog Biophys Mol Biol. 2006;92(1):60–4. doi: 10.1016/j.pbiomolbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 63.Froicu M, Cantorna MT. Vitamin D and the vitamin D receptor are critical for control of the innate immune response to colonic injury. BMC Immunol. 2007;8:5. doi: 10.1186/1471-2172-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Froicu M, Weaver V, Wynn TA, et al. A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol. 2003;17(12):2386–92. doi: 10.1210/me.2003-0281. [DOI] [PubMed] [Google Scholar]
- 65.Froicu M, Zhu Y, Cantorna MT. Vitamin D receptor is required to control gastrointestinal immunity in IL-10 knockout mice. Immunology. 2006;117(3):310–8. doi: 10.1111/j.1365-2567.2005.02290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong J, Zhang Z, Musch MW, et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008;294(1):G208–16. doi: 10.1152/ajpgi.00398.2007. [DOI] [PubMed] [Google Scholar]
- 67.Wu S, Sun J. Vitamin D, vitamin D receptor, and macroautophagy in inflammation and infection. Discov Med. 2011;11(59):325–35. [PMC free article] [PubMed] [Google Scholar]
- 68.Wang TT, Dabbas B, Laperriere D, et al. Direct and indirect induction by 1,25-dihydroxyvitamin D3 of the NOD2/CARD15-defensin beta2 innate immune pathway defective in Crohn disease. J Biol Chem. 2010;285(4):2227–31. doi: 10.1074/jbc.C109.071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramagopalan SV, Heger A, Berlanga AJ, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20(10):1352–60. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleming A, Noda T, Yoshimori T, et al. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7(1):9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 71.Balgi AD, Fonseca BD, Donohue E, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4(9):e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104(48):19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singletary K, Milner J. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1596–610. doi: 10.1158/1055-9965.EPI-07-2917. [DOI] [PubMed] [Google Scholar]
- 74.Matsuda C, Ito T, Song J, et al. Therapeutic effect of a new immunosuppressive agent, everolimus, on interleukin-10 gene-deficient mice with colitis. Clin Exp Immunol. 2007;148(2):348–59. doi: 10.1111/j.1365-2249.2007.03345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dumortier J, Lapalus MG, Guillaud O, et al. Everolimus for refractory Crohn's disease: a case report. Inflamm Bowel Dis. 2008;14(6):874–7. doi: 10.1002/ibd.20395. [DOI] [PubMed] [Google Scholar]
- 76.Massey DC, Bredin F, Parkes M. Use of sirolimus (rapamycin) to treat refractory Crohn's disease. Gut. 2008;57(9):1294–6. doi: 10.1136/gut.2008.157297. [DOI] [PubMed] [Google Scholar]
- 77.Reinisch W, Panes J, Lemann M, et al. A multicenter, randomized, double-blind trial of everolimus versus azathioprine and placebo to maintain steroid-induced remission in patients with moderate-to-severe active Crohn's disease. Am J Gastroenterol. 2008;103(9):2284–92. doi: 10.1111/j.1572-0241.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 78.Cantorna MT, Munsick C, Bemiss C, et al. 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease. J Nutr. 2000;130(11):2648–52. doi: 10.1093/jn/130.11.2648. [DOI] [PubMed] [Google Scholar]
- 79.Laverny G, Penna G, Vetrano S, et al. Efficacy of a potent and safe vitamin D receptor agonist for the treatment of inflammatory bowel disease. Immunol Lett. 2010;131(1):49–58. doi: 10.1016/j.imlet.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 80.Lagishetty V, Misharin AV, Liu NQ, et al. Vitamin D deficiency in mice impairs colonic antibacterial activity and predisposes to colitis. Endocrinology. 2010;151(6):2423–32. doi: 10.1210/en.2010-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miheller P, Muzes G, Hritz I, et al. Comparison of the effects of 1,25 dihydroxyvitamin D and 25 hydroxyvitamin D on bone pathology and disease activity in Crohn's disease patients. Inflamm Bowel Dis. 2009;15(11):1656–62. doi: 10.1002/ibd.20947. [DOI] [PubMed] [Google Scholar]
- 82.Jorgensen SP, Agnholt J, Glerup H, et al. Clinical trial: vitamin D3 treatment in Crohn's disease - a randomized double-blind placebo-controlled study. Aliment Pharmacol Ther. 2010;32(3):377–83. doi: 10.1111/j.1365-2036.2010.04355.x. [DOI] [PubMed] [Google Scholar]