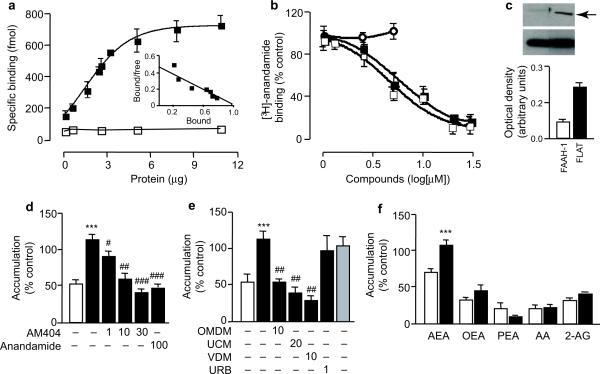
Fig. 2.
FLAT binds to anandamide and facilitates its transport into cells. (a) Specific binding of [3H]-anandamide to rat FLAT-glutathione-S-transferase (GST) (closed squares) or GST alone (open squares). The inset shows a Scatchard transformation (bound - [bound/free], in nmol) of binding data. (b) AM404 (closed squares) and OMDM-1 (closed squares) antagonize [3H]-anandamide binding to FLAT-GST, whereas URB597 (open circles) has no effect. (c) Cytosolic fractions of FLAT-expressing Hek293 cells contain detectable amounts of FLAT (arrow); a corresponding fraction from FAAH-1-expressing cells is shown for comparison. The lower band is β-actin. (d) [3H]- Anandamide accumulation in control cells (vector-transfected, open bar) or FLAT-expressing Hek293 cells (closed bars) incubated with vehicle (0.01%dimethylsulfoxide), AM404 or non-radioactive anandamide (concentrations in μM). (e) The anandamide transport inhibitors OMDM-1, UCM-707 and VDM-11 suppress [3H]-anandamide accumulation in FLAT-expressing Hek293 cells. This process is not affected by URB597 or mutation of catalytic Ser242 (shaded bar). (f) Accumulation of labeled lipids in control (open bars) or FLAT-expressing Hek293 cells (closed bars). Abbreviations: [3H]-anandamide, AEA; [3H]-oleoylethanolamide, OEA; [3H]-palmitoylethanolamide, PEA; [3H]-arachidonic acid, AA; [3H]-2-arachidonoyl-sn-glycerol, 2-AG. Results are expressed as mean±SEM of 3-7 experiments. ***P<0.01, versus vector-transfected cells, Student's t test; #P<0.05; ##, P<0.01; ###, P<0.001 versus vehicle, one-way ANOVA followed by Dunnett's test.