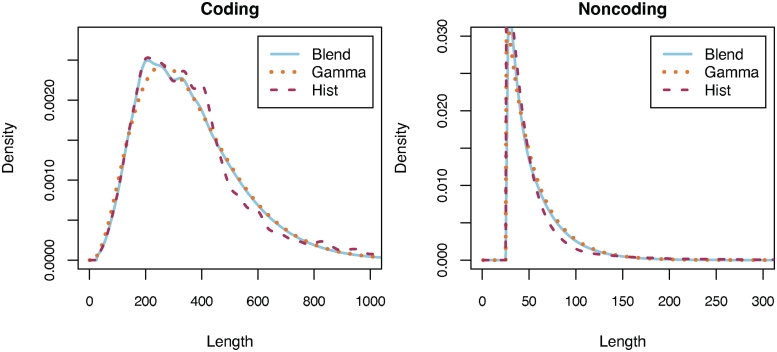
Figure 1.
Distributions for coding and non-coding ORF lengths (in amino acids) from Deinococcus radiodurans R1 estimated using the Gamma distribution (Gamma), a smoothed histogram (Hist), and a blend of the first two (Blend) that uses the histogram model for the first quartile, the Gamma model for the last quartile, and a linear combination in between. The Hist model offers greater accuracy for short and medium sized ORFs (e.g. the deviation from Gamma at 200 bp in the coding plot), but is useless for very long ORFs, which Gamma can model more effectively. The shape of the D. radiodurans length distributions are typical of the prokaryotic genomes examined, but Glimmer-MG estimates the distributions for each genome individually.