Abstract
In eukaryotes, the cytoplasmic and mitochondrial forms of a given aminoacyl-tRNA synthetase (aaRS) are typically encoded by two orthologous nuclear genes, one of eukaryotic origin and the other of mitochondrial origin. We herein report a novel scenario of aaRS evolution in yeast. While all other yeast species studied possess a single nuclear gene encoding both forms of alanyl-tRNA synthetase (AlaRS), Vanderwaltozyma polyspora, a yeast species descended from the same whole-genome duplication event as Saccharomyces cerevisiae, contains two distinct nuclear AlaRS genes, one specifying the cytoplasmic form and the other its mitochondrial counterpart. The protein sequences of these two isoforms are very similar to each other. The isoforms are actively expressed in vivo and are exclusively localized in their respective cellular compartments. Despite the presence of a promising AUG initiator candidate, the gene encoding the mitochondrial form is actually initiated from upstream non-AUG codons. A phylogenetic analysis further revealed that all yeast AlaRS genes, including those in V. polyspora, are of mitochondrial origin. These findings underscore the possibility that contemporary AlaRS genes in V. polyspora arose relatively recently from duplication of a dual-functional predecessor of mitochondrial origin.
INTRODUCTION
Faithful decoding of mRNA into protein depends on accurate aminoacylation of tRNA by aminoacyl-tRNA synthetases (aaRSs) and specific codon/anticodon base pairing. AaRSs are a group of structurally diverse enzymes responsible for protein translation, each of which couples a specific amino acid to its cognate tRNA. The resultant aminoacyl-tRNAs are then delivered by elongation factor (EF)-1 to ribosomes for protein synthesis. In prokaryotes, there are typically 20 aaRSs, one for each amino acid (1–3). In eukaryotes, protein synthesis occurs in the cytoplasm and organelles, such as mitochondria and chloroplasts (4). Thus, eukaryotes, such as yeast, commonly have two genes that encode distinct sets of proteins for each aminoacylation activity, one localized in the cytoplasm and the other in mitochondria. Each set aminoacylates the tRNAs within its respective cellular compartment, and is sequestered from isoacceptors confined to other cellular compartments.
Cytoplasmic and mitochondrial forms of an aaRS are commonly encoded by two nuclear genes of distinct evolutionary origins in eukaryotes (5). Typically, the gene that encodes the cytoplasmic form is of eukaryotic (or archaeal) origin, while the gene that encodes its mitochondrial counterpart is of prokaryotic origin. However, in some cases, a single nuclear gene encodes both forms of a given aaRS through alternative initiation of translation, while its orthologue was lost during evolution. Examples of this type include yeast ALA1 [which encodes alanyl-tRNA synthetase (AlaRS)] (6), GRS1 (which encodes glycyl-tRNA synthetase) (7), HTS1 (which encodes histidyl-tRNA synthetase) (8) and VAS1 (which encodes valyl-tRNA synthetase) (9). Since isoforms target different cellular compartments, they cannot be substituted for one another in vivo. A similar phenotype was observed for genes encoding cytoplasmic and mitochondrial forms of Arabidopsis thaliana AlaRS, threonyl-tRNA synthetase and valyl-tRNA synthetase (10). With the exception of a few controversial incidents caused by genetic recombinations or horizontal gene transfer, the evolutionary scenarios of eukaryotic aaRSs largely fit in one of the two paradigms described above. Nevertheless, it was recently reported that two paralogous valyl-tRNA synthetase genes of mitochondrial origin coexist in the fission yeast Schizosaccharomyces pome, one that encodes the cytoplasmic form and the other its mitochondrial counterpart (11).
Many examples of non-AUG initiation were reported in higher eukaryotes, where cellular and viral mRNAs initiate from codons that differ from AUG by one nucleotide, such as ACG, UUG and AUU (12). However, only a few cases were found to date in low eukaryotes such as yeast. ALA1 and GRS1 are among the first genes shown to use naturally occurring non-AUG triplets as an alternative translation initiator in the model yeast S. cerevisiae (6,7). While sequence context has little effect on the translational efficiency of AUG initiation in yeast (13–15), translation initiation from a non-AUG codon is profoundly affected (by up to 32-fold) by nucleotides at relative positions −3 to −1, especially −3. AARuug (R denotes A or G; uug denotes a non-AUG initiation codon) appears to represent the most favorable sequence context (16,17). An interesting feature of the expression of ALA1 is that this gene contains two consecutive ACG initiator codons for its mitochondrial form (6). Consecutive ACG codons possess stronger initiation activity than does a single ACG codon (18). This feature of remedial initiation from repeated non-AUG initiator codons per se may represent a novel mechanism to improve the efficiency of translation initiation. Examples of non-AUG initiation were also recently reported in other yeast species (11,19).
As aaRSs are one of the most ancient groups of enzymes that continue to function in all free-living organisms, they are often targeted for taxonomic and phylogenetic studies (20–22). In this report, we explored the functions and historical relationships of AlaRSs from 13 different yeast species. It is our hope that results obtained in this study will advance our understanding of the biochemical properties of the eukaryotic aaRS gene, and provide new insights into its evolutionary footprints. Results presented herein show that while all other yeast species studied possess a single AlaRS gene with dual functions, Vanderwaltozyma polyspora, a close relative of S. cerevisiae, contains two distinct AlaRS genes that respectively encode the cytoplasmic and mitochondrial forms. Unexpectedly, all yeast AlaRS genes are of the mitochondrial type, irrespective of the cellular compartments where the protein products are active. These and other results imply that AlaRS genes in V. polyspora underwent a major genetic recombination at least twice. First, the mitochondrion-type gene gained cytoplasmic function and became a dual-functional gene, while its orthologue was lost. Later, the dual-functional gene of mitochondrial origin was duplicated into two copies, each retaining a single function (cytoplasmic or mitochondrial).
MATERIALS AND METHODS
Construction of plasmids
Cloning of ALA1 genes of S. cerevisiae and Candida albicans was previously described (6,23). To clone the ALA1 gene of V. polyspora (VpALA1) into pRS315-2xFLAG (a low-copy-number yeast shuttle vector carrying a LEU2 marker and a 2xFLAG sequence), a pair of gene-specific primers was used to amplify the gene via a polymerase chain reaction (PCR) using yeast genomic DNA as the template. The forward primer with an EagI site is located 300 bp upstream of the first ATG codon of the open reading frame (ORF), while the reverse primer with an XhoI site is located immediately upstream of the stop codon. The ~3.3-kb PCR-amplified DNA fragment was subsequently digested with EagI and XhoI prior to cloning into pRS315. Cloning of VpALA2 and SpALA1 into pRS315 followed a similar protocol. Note that these clones were expressed under the control of their respective native promoters.
Cloning of SpALA1 into pADH (a high-copy-number yeast shuttle vector with a LEU2 marker, an ADH promoter preceding the multiple cloning sites, and a 2xFLAG sequence following the multiple cloning sites) (24) followed a similar protocol, except that the forward primer was annealed to a sequence 12 bp upstream of the first ATG codon. Note that this clone was expressed under the control of a constitutive ADH promoter. Construction of initiator mutants of VpALA1 and VpALA2 followed a strategy described earlier (7). Mutagenesis was carried out following a protocol provided by the manufacturer (Stratagene, La Jolla, CA, USA). Green fluorescence protein (GFP) assays followed a protocol described elsewhere (25).
Complementation assays for cytoplasmic AlaRS activity
The yeast ALA1 knockout strain, TRY11 (MATa, his3Δ200, leu2Δ1, lys2-801, trp1Δ101, ura3-52 and ala1Δ::TRP1) was maintained by a plasmid carrying the wild-type ALA1 gene and a URA3 marker (26). Complementation assays of cytoplasmic AlaRS activity were carried out by introducing a test plasmid carrying the gene of interest and a LEU2 marker into TRY11, and the ability of the transformants to grow in the presence of 5-FOA was determined. Starting from a cell density of 4.0 A600, cell cultures were 5-fold serially diluted, and 10 -μl aliquots of each dilution were spotted onto designated plates containing 5-FOA. Plates were incubated at 30°C for 3–5 days. The transformants evicted the maintenance plasmid with a URA3 marker in the presence of 5-FOA, and thus could not grow on the selection medium unless a functional cytoplasmic AlaRS was encoded by the test plasmid.
Complementation assays for mitochondrial AlaRS activity
TRY11 was co-transformed with a test plasmid and a second maintenance plasmid (carrying a HIS3 marker) that only expresses the cytoplasmic form of AlaRS (due to a mutation in the non-AUG initiator codon). In the presence of 5-FOA, the first maintenance plasmid (carrying a URA3 marker) was evicted from the co-transformants, while the second maintenance plasmid was retained. Thus, all co-transformants survived 5-FOA selection due to the presence of the cytoplasmic AlaRS derived from the second maintenance plasmid. The mitochondrial phenotypes of the co-transformants were further tested on YPG plates at 30 C, with results documented on Day 3 following plating. Because a yeast cell cannot survive on glycerol without functional mitochondria, the co-transformants did not grow on YPG plates unless a functional mitochondrial AlaRS was generated from the test plasmid.
Reverse transcription—PCR
To determine the relative levels of endogenous ALA1 and ALA2 mRNAs in V. polyspora, a semiquantitative reverse transcription (RT)–PCR experiment was carried out following the protocols provided by the manufacturer (Invitrogen, Carlsbad, CA, USA). Total RNA was first isolated from the transformant, and then treated with DNase to remove contaminating DNA. Aliquots (~1 µg) of the RNA were then reverse-transcribed into single-stranded complementary (c)DNA using an oligo-dT primer. After RNase H treatment, single-stranded cDNA products were amplified by a PCR using a pair of gene-specific primers. The forward and reverse primers (GSP1 and GSP2) for VpALA1 contained sequences complementary to nucleotides 1936–1961 (5′-CGTAATGTCGAAGAAGTCTGTAACC-3′) and nucleotides 2379–2405 (5′-TAATGACAGAAATGGATAGTTGACC-3′) of VpALA1, respectively. The forward and reverse primers (GSP3 and G-SP4) for VpALA2 contained sequences complementary to nucleotides 1966–1992 (5′-TTGATTGTCAGTAAGAAACCTGTC-3′) and nucleotides 2320–2346 (5′-CGTCAGAAAATGGTAGTGAGTTCAG-3′) of VpALA2, respectively. To obtain more-reliable data, four different cycle numbers of PCR were carried out for each cDNA preparation as indicated in the figure.
A quantitative real-time RT–PCR experiment was subsequently used to obtain more-accurate data. Two sets of primers, GSP5/GSP6 and GSP7/GSP8, were respectively used to amplify the cDNA fragments of VpALA1 and VpALA2. GSP5 and GSP6 are complementary to nucleotides +1998 to +2022 and +2111 to +2135 of VpALA1, respectively, while GSP7 and GSP8 are complementary to nucleotides +2004 to +2028 and +2016 to +2040 of VpALA2, respectively. Quantitative data were obtained from three independent experiments and averaged.
Western blot analysis
Protein expression patterns of the ALA1 and ALA2 constructs were determined by a chemiluminescence-based western blot analysis. INVSc1 (MATa, his3Δ1, leu2, trp1-289, ura3-52; MATα, his3Δ1, leu2, trp1-289, ura3-52) (Invitrogen) was first transformed with the constructs of interest, and total protein extracts were prepared from the transformants. Aliquots of protein extracts (40 μg) were loaded onto a mini gel (of 8 × 10 cm) containing 10% polyacrylamide and electrophoresed at 100 V for 1–2 h. Following electrophoresis, the resolved proteins were transferred using a semi-dry transfer device to a polyvinylidene fluoride (PVDF) membrane in buffer containing 30 mM glycine, 48 mM Tris base (pH 8.3), 0.037% sodium dodecylsulfate (SDS) and 20% methanol. The membrane was probed with an anti-FLAG tag antibody (Sigma) followed by an HRP-conjugated goat anti-mouse IgG antibody (Invitrogen), and then exposed to X-ray film following the addition of appropriate substrates.
RESULTS
Vanderwaltozyma polyspora possesses two distinct nuclear AlaRS genes
Previous studies showed that the ALA1 gene of S. cerevisiae encodes both mitochondrial and cytoplasmic forms of AlaRS (6,23). We wondered whether a similar feature was conserved in the AlaRS genes of other yeast species. Pursuant to this objective, 14 different yeast AlaRS genes were retrieved from the EMBL database, and the predicted amino acid sequences were analyzed. Except for V. polyspora, which contained two distinct nuclear AlaRS genes (designated herein as VpALA1 and VpALA2), all yeast species studied possessed a single AlaRS gene (designated herein as ALA1) (Figure 1). As aminoacylation activities are required for protein synthesis in both the cytoplasm and mitochondria, the single AlaRS gene was expected to provide both cytoplasmic and mitochondrial functions in these yeast species. Indeed, two in-frame initiator candidates were found in each of the ORFs (Figure 1A). Despite only one ATG initiator codon being found in ALA1 genes of Ashbya gossypii, C. glabrata and Kluyveromyces lactis, a promising non-ATG initiator candidate was mapped in the leader sequence of these genes (Figure 1A). In agreement with this tentative assignment, the N-terminal pre-sequences deduced from these genes were all rich in positively charged and hydroxylated amino acid residues but were devoid of acidic residues, characteristic of a mitochondrial targeting signal.
Figure 1.
Alignment of N-terminal presequences of yeast AlaRSs. (A) N-terminal pre-sequences of yeast AlaRSs. Initiating methionines for the cytoplasmic and mitochondrial forms of AlaRS are in boldface. The letter ‘m’ denotes putative initiation from a non-AUG codon. Sc, Saccharomyces cerevisiae (NP_014980); Ca, Candida albicans (XP_718016); Yl, Yarrowia lipolytica (XP_500483); Sp, S. pombe (NP_593640); Ag, Ashbya gossypii (NP_984459); Cg, Candida glabrata (XP_445923); Cn, Cryptococcus neoformans (XP_774788); Dh, Debaryomyces hansenii (XP_456501); Kl, Kluyveromyces lactis (XP_455190); Mg, Malassezia globosa (XP_001731546); Pg, Pichia guilliermondii (XP_001486846); Ps, Pichia stipitis (XP_001383420); and Vp, Vanderwaltozyma polyspora (VpAlaRS1, XP_001643652; VpAlaRS2, XP_001642960). (B) 5′-End sequences of VpALA1 and VpALA2. The sequence extends from the nearest upstream stop codon to +60 nucleotide relative to ATG1. ATG and potential non-ATG initiator codons are underlined. (C) Sequence homology among ScAlaRS, VpAlaRS1 and VpAlaRS2.
Despite V. polyspora containing two distinct nuclear AlaRS genes, neither of the protein forms deduced from the ORFs of these two genes (starting from the ATG1 codon) possessed a canonical N-terminal mitochondrial targeting signal. Sequence alignment further indicated that the first ATG codons of these two genes were both located at a position comparable to that of the ATG initiator codon for the cytoplasmic form of ScAlaRS (AlaRS of S. cerevisiae) (Figure 1A). A closer look at the 5′-end sequences of these two genes suggested that ATG1 was the only possible initiator codon for VpALA1, while VpALA2 contained several promising non-ATG initiator candidates 5′ to ATG1, such as TTG(−28), ACG(−23) and TTG(−15) (Figure 1B). The protein forms deduced from the ORFs of these two genes considerably resembled each other (~66% identity), with VpAlaRS1 (AlaRS1 of V. polyspora) having a higher similarity to ScAlaRS (83% identity) (Figure 1C).
The top three base pairs in the acceptor stem are highly conserved in all yeast tRNAsAla
Previous studies showed that the major identity elements of tRNAAla reside in its acceptor stem. In particular, a G3:U70 base pair is conserved in the acceptor stems of all known tRNAsAla from prokaryotes, archaea, eukaryote cytoplasm and chloroplasts (27,28). Comparison of acceptor stems of tRNAsAla from various yeast species indicated that G3:U70, G1:C72 and G2:C71 are strictly conserved in the acceptor stems of all yeast tRNAAla isoacceptors, irrespective of whether they are nuclear- or mitochondrion-encoded tRNAs (Figure 2). In contrast to the stringent conservation in the top three base pairs of the acceptor stem, the bottom four base pairs appeared relatively divergent, in particular between the cytoplasmic and mitochondrial tRNAAla molecules. Cytoplasmic tRNAsAla strongly preferred C4:G69, G5:C68, U6:U67 and G7:C66, while the mitochondrial counterparts moderately preferred G4:C69, U5:A68, U6:A67 and A7:U66. This preference for particular base pairs in the bottom part of the acceptor stem of yeast tRNAAla isoacceptors may reflect differences in their evolutionary origins. From this perspective, it was interesting to note that while V. polyspora possessed two distinct AlaRS genes, the acceptor stems of their cognate tRNAs showed a preference similar to that of other yeast tRNAsAla (Figure 2).
Figure 2.
Comparison of the acceptor stems of yeast tRNAAla isoacceptors. Acceptor stems of representative cytoplasmic (upper panel) and mitochondrial (lower panel) tRNAsAla from various yeast species are shown for comparison. The primary identity element, G3:U70, for tRNAAla is marked with a short line. cyt, cytoplasmic; mit, mitochondrial.
Vanderwaltozyma polyspora ALA1 and ALA2 encode the cytoplasmic and mitochondrial forms of AlaRS, respectively
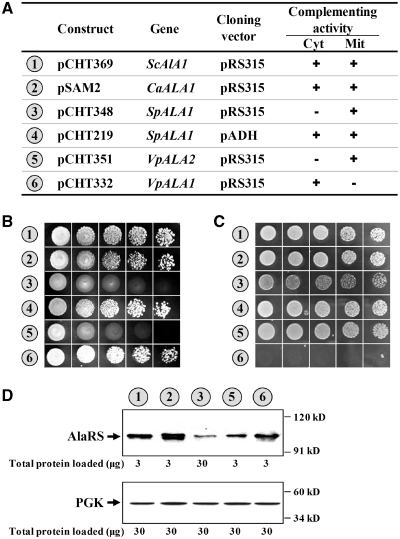
To further investigate the functional potential of these AlaRS genes, several representative sequences were cloned in pRS315 (a low-copy-number yeast shuttle vector), and their cross-species complementation activities were assayed in TRY11, an ala1- strain of S. cerevisiae. Consistent with previous reports, CaALA1 and ScALA1 rescued both the cytoplasmic and mitochondrial defects of the knockout strain (6,23). Transformants bearing either of these two constructs grew robustly on 5-FOA and YPG plates (Figure 3A–C, nos 1 and 2). In contrast, SpALA1, when cloned in the same vector―pRS315, rescued only the mitochondrial defect of the knockout strain (Figure 3A–C, no. 3), presumably due to poor gene expression by its native promoter (Figure 3D). A dual-functional phenotype was observed for this gene when it was cloned into pADH, a high-copy number yeast shuttle vector with a constitutive ADH promoter (Figure 3A–C, no. 4). Thus, SpAlaRS can recognize both cytoplasmic and mitochondrial tRNAAla isoacceptors of S. cerevisiae. In contrast to the dual-functional phenotype of these genes, AlaRS genes from V. polyspora appeared to have single functions. VpALA1 and VpALA2, respectively, rescued the cytoplasmic and mitochondrial defects of TRY11, suggesting that these two genes respectively provide cytoplasmic and mitochondrial functions (Figure 3B and C, nos 5 and 6). The protein expression patterns of these genes were subsequently determined by western blotting. As shown in Figure 3D, all genes, except for SpALA1, were well expressed from their native promoters in S. cerevisiae. It appeared that the native promoter of SpALA1 cannot efficiently be recognized by the transcription apparatus of S. cerevisiae. In addition, the protein expression level of VpALA2 was much lower than that of VpALA1.
Figure 3.
Cross-species complementation assays for various yeast AlaRS genes. Various ALA1 constructs were transformed into TRY11, an ala1− strain of S. cerevisiae, and the ability of the constructs to rescue the growth defects of the knockout strain was tested. (A) Schematic summary of the ALA1 and ALA2 constructs and their complementation activities. The symbols ‘+’ and ‘−’ denote positive and negative complementation, respectively. mit, mitochondrial; cyt, cytoplasmic. (B) Complementation assays for cytoplasmic AlaRS activity. (C) Complementation assays for mitochondrial AlaRS activity. Numbers 1–8 (circled) in (A–C) denote the constructs shown in (A). (D) Western blotting using an anti-FLAG antibody. Upper panel, AlaRS; lower panel, PGK (as a loading control).
Vanderwaltozyma polyspora ALA2 is exclusively initiated from upstream non-AUG codons
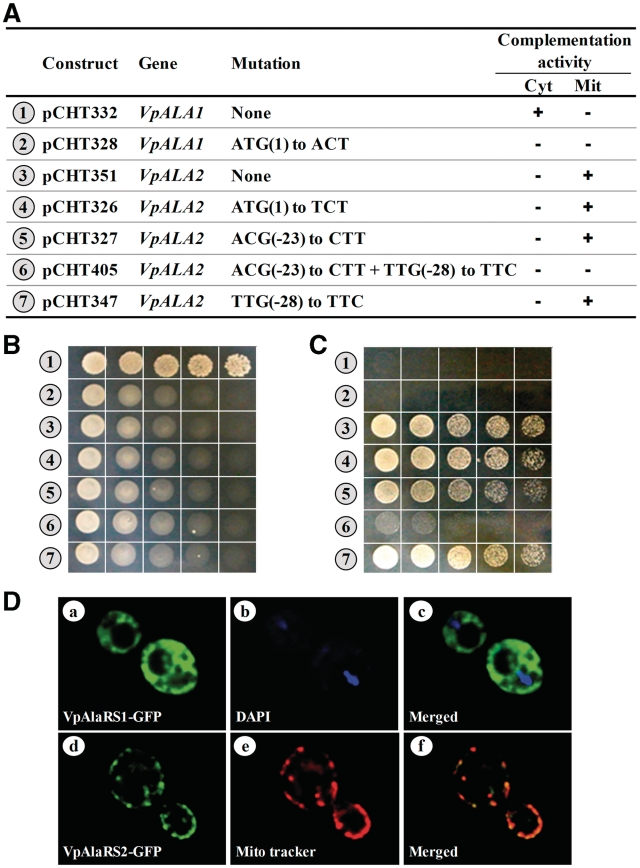
As mentioned above, no N-terminal mitochondrial targeting signal was predicted for the ATG1-initiated protein sequence of VpAlaRS2. How is this protein form targeted to mitochondria for its functioning? To verify whether this gene is actually initiated from ATG1, this codon was subsequently mutated, and the complementation activity of the resultant construct was tested. As a reference, the first ATG codon of VpALA1 was also mutated and tested. As expected, a mutation at ATG1 of VpALA1 drastically impaired its cytoplasmic activity. However, a mutation at ATG1 of VpALA2 had little effect on its mitochondrial activity (Figure 4A–C, nos 2 and 4). This result suggests that VpALA1 is initiated from ATG1, while VpALA2 is initiated elsewhere.
Figure 4.
Mapping of the translation initiator codons of VpALA1 and VpALA2. Wild-type and mutant ALA1 or ALA2 constructs were transformed into TRY11, and the ability of the constructs to rescue growth defects of the knockout strain was tested. (A) Schematic summary of the ALA1 and ALA2 constructs and their complementation activities. (B) Complementation assays for cytoplasmic AlaRS activity. (C) Complementation assays for mitochondrial AlaRS activity. (D) Analysis of cellular localization of VpAlaRS1 and VpAlaRS2 by fluorescence microscopy. A mitochondrial tracker and DAPI were used to label mitochondria and nuclei, respectively.
To obtain more clues into solving this mystery, upstream in-frame non-ATG codons that fit the following three criteria were preferentially selected as initiator candidates and tested for their initiating activity. First, they had to differ from ATG by only one nucleotide. Second, they had to bear an ‘A’ nucleotide at relative position −3. Lastly, they had to possess the potential to initiate translation of a mitochondrial targeting signal devoid of acidic amino acid residues. The only candidate that perfectly matched all of these criteria was ACG(−23). Unexpectedly, a mutation at this candidate slightly reduced, but not eliminated, the mitochondrial activity of VpALA2 (Figure 4A–C, no. 5), suggesting the presence of an alternative initiation site. To gain further insights, a less likely candidate, TTG(−28), that matched some of the criteria described above was also tested for its initiation activity. While a mutation at TTG(−28) alone had little effect on the mitochondrial activity of VpALA2, simultaneous mutations at ACG(−23) and TTG(−28) drastically impaired its mitochondrial activity (Figure 4A–C, nos. 6–7). Note that TTG(−28) contained a ‘C’ at relative position −3. Mainly for this reason, TTG(−28) was not selected as one of the primary targets in the first round. So, despite the presence of a promising ATG initiator candidate, VpALA2 was exclusively initiated from upstream non-ATG codons, with ACG(−23) being the major initiation site and TTG(−28) being the minor initiation site.
To verify the cellular localization of these two proteins in vivo, a DNA sequence encoding the GFP was inserted in-frame at the 3′-ends of these two genes, resulting in VpALA1-GFP and VpALA2-GFP. As genetic tools for V. polyspora are not yet fully developed, we herein used an S. cerevisiae strain, INVScI, as the platform for the localization assay. The GFP fusion constructs were individually transformed into INVSc1, and the subcellular localization of these GFP fusion proteins in the transformants was examined under fluorescence microscopy. As shown in Figure 4D, VpAlaRS2-GFP exclusively existed in mitochondria (d–f), while VpAlaRS1-GFP was evenly distributed in the cytosol (a–c). This result provides further support to the notion that VpALA1 and VpALA2 encode the cytoplasmic and mitochondrial forms of AlaRS, respectively.
Both ALA1 and ALA2 are actively expressed in Vanderwaltozyma polyspora
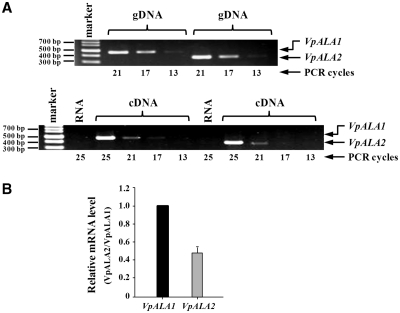
To investigate whether VpALA1 and VpALA2 are actively expressed in their native host, V. polyspora, relative levels of their specific mRNAs were determined by a semiquantitative RT-PCR. Two sets of gene-specific primers were designed, one set complementary to VpALA1 and the other to VpALA2. The expected sizes of the PCR-amplified products were 469 and 380 base pairs for VpALA1 and VpALA2, respectively. As shown in Figure 5A, similar levels of VpALA1 and VpALA2 DNA fragments were amplified by their respective primers using the genomic DNA of V. polyspora as a template (upper panel), suggesting that these two sets of primers were annealed to their respective target genes with similar efficiencies under the conditions used. The same sets of primers were subsequently used to amplify the cDNA of these two genes, which had been reverse-transcribed from a total RNA preparation of V. polyspora using oligo-dT as the primer. Figure 5A shows that similar levels of VpALA1 and VpALA2 DNA fragments were amplified using the reverse-transcribed cDNA as a template (lower panel), suggesting that both genes are actively expressed in V. polyspora and are transcribed at comparable efficiencies. Further determination using a quantitative (q)RT–PCR shows that VpALA1 had a transcriptional efficiency two-fold higher than that of VpALA2 (Figure 5B). Note that these two sets of primers were annealed to their respective target genes with an almost identical efficiency using the genomic DNA of V. polyspora as a template.
Figure 5.
Analysis of endogenous VpALA1 and VpALA2 mRNAs by RT–PCR. (A) A semiquantitative RT–PCR. PCR amplification of VpALA1 and VpALA2 DNA fragments using genomic (g)DNA as the template (upper panel); RT–PCR amplification of VpALA1 and VpALA2 mRNAs using complementary (c)DNA as the template (lower panel). RNA control denotes PCR amplification using RNA, instead of cDNA, as the template. (B) qRT–PCR.
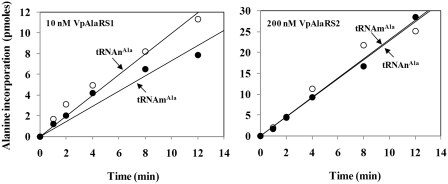
To test the substrate specificities of VpAlaRS1 and VpAlaRS2, recombinant AlaRS-His6 enzymes were purified from yeast transformants harboring appropriate plasmids by Ni-NTA column chromatography to homogeneity, and in vitro-transcribed tRNAnAla (nuclear-encoded cytosolic tRNAAla) and tRNAmAla (mitochondrion-encoded tRNAAla) of V. polyspora were used as the substrates for the aminoacylation reactions. Figure 6 shows that VpAlaRS1 and VpAlaRS2 had a similar tRNA specificity. This result reinforces the hypothesis that localization of these two enzymes determines their functions in vivo.
Figure 6.
Aminoacylation activities of VpAlaRS1 and VpAlaRS2. Aminoacylation activities of purified recombinant AlaRS-His6 enzymes were determined at 30°C by measuring the relative amounts of 3H-alanine that were incorporated into tRNA using a liquid scintillation counter (11). Various tRNAs were prepared by in vitro transcription. tRNAnAla and tRNAmAla represent cytoplasmic and mitochondrial tRNAAla isoacceptors, respectively.
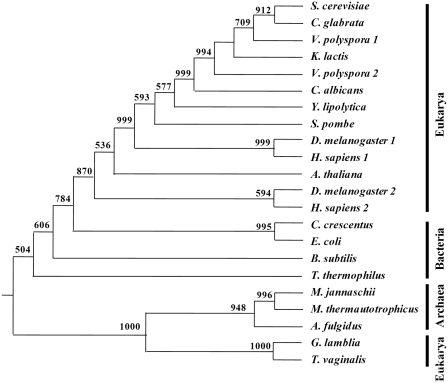
Both ALA1 and ALA2 of Vanderwaltozyma polyspora are of mitochondrial origin
As V. polyspora is so far the only known yeast species that possesses two distinct AlaRS genes (Figure 1), possible historical relationships between VpAlaRS1, VpAlaRS2 and their yeast homologues were analyzed using the neighbor-joining method. To better illustrate this phylogeny, representative AlaRS sequences from all three of the major domains of life (Bacteria, Archaea and Eukarya) were also subjected to the analysis. Bias in the alignments of these sequences was eliminated by reducing them to their core active sites. This portion comprises only ~53% of the sequence of V. polyspora AlaRS2. As shown in Figure 7, except for AlaRS sequences from mitochondrion-deficient eukaryotes Giardia lamblia and Trichomonas vaginalis, all eukaryotic AlaRS sequences, including those from S. cerevisiae, C. albicans, Y. lipolytica, V. polyspora, A. thaliana, Drosophila melanogaster (cytoplasmic and mitochondrial isoforms) and Homo sapiens (cytoplasmic and mitochondrial isoforms), clustered within a monophyletic branch that showed a higher similarity to the bacterial branch than to the archaeal branch (Figure 7). This finding suggests that all AlaRS sequences from mitochondrion-containing eukaryotes, including those from yeasts, are of the mitochondrial type. Thus, irrespective of the cellular compartments in which their protein products are active, VpALA1 and VpALA2 actually arose from the same ancestor. Moreover, it appears that loss of the AlaRS gene of eukaryotic origin in mitochondrion-containing eukaryotes occurred long before the events of yeast speciation.
Figure 7.
Phylogenetic analysis of the relationships of VpAlaRS1, VpAlaRS2 and other AlaRSs. Sequences comprising the core active sites of AlaRSs were aligned with Clustal W (38) and analyzed using the Neighbor-joining method. Numbers at the nodes denote bootstrapping frequencies calculated from 1000 trees. A. fulgidus, Archaeoglobus fulgidus (NP_071080); A. thaliana, Arabidopsis thaliana (CAA80380); B. subtilis, Bacillus subtilis (NP_390618); C. crescentus, Caulobacter crescentus (NP_421332); D. melanogaster, Drosophila melanogaster (DmAlaRS1, NP_523511.2; DmAlaRS2, NP_523932.2); E. coli, Escherichia coli (NP_417177); G. lamblia, Giardia lamblia (XP_001704452); H. sapiens, Homo sapiens (HsAlaRS1, BAA06808; HsAlaRS2, NP_065796); M. jannaschii, Methanocaldococcus jannaschii (NP_247543); M. thermautotrophicus, Methanothermobacter thermautotrophicus (NP_276794); T. thermophilus, Thermus thermophilus (YP_145097); T. vaginalis, Trichomonas vaginalis (AAT95880.1).
DISCUSSION
As aaRSs are likely to be one of the most ancient families of proteins, they are often targets of choice for phylogenetic analyses. Results presented herein showed that while all other yeasts contained a single AlaRS gene with dual functions, V. polyspora possessed two distinct AlaRS genes (VpALA1 and VpALA2) (Figures 1 and 3). VpALA1 and VpALA2 can substitute for the cytoplasmic and mitochondrial functions of S. cerevisiae ALA1, respectively (Figures 3 and 4). Strangely enough, the protein sequence deduced from the ORF of VpALA2 (starting from ATG1) lacks a cleavable N-terminal mitochondrial targeting signal as predicted by a PSORTII analysis (29). In fact, the ATG1 codon of this gene is located at a position equivalent to that of the ATG initiator codon for the cytoplasmic form of S. cerevisiae AlaRS (Figure 1). Further investigation revealed that VpALA2 is exclusively initiated from upstream non-ATG codons (Figure 4). This unexpected finding may account for the observation that while this gene is transcribed at an efficiency equivalent to that of its counterpart (i.e. VpALA1) (Figure 5), it has a much lower protein expression level (Figure 3). Also unexpected is the finding that one of the non-ATG initiator codons of VpALA2 contains a less favorable nucleotide, C, at relative position -3 (Figures 1 and 4). Unlike most other examples of alternative initiation of translation (30–33), the downstream ATG initiator candidate of VpALA2 appears to be silent in this case.
The specificity of an aminoacylation reaction is generally accomplished by direct recognition of tRNA by its cognate aaRS. In some instances, recognition depends mainly on the anticodon loop, while in others it relies more on sequences or structures in the acceptor stem (34). In the instance of tRNAAla, the primary identity determinant is a G3:U70 base pair, which is strictly conserved in the acceptor stems of all known tRNAAla molecules from prokaryotes, archaea, eukaryote cytoplasm and chloroplasts (27,28). This strong preference for a single determinant may provide a functional basis for the high feasibility and high efficiency of cross-species and cross-compartmental complementation (Figures 3 and 4) (6). Amazingly, even a human AlaRS gene can complement an ALA1 knockout strain of S. cerevisiae (26). On the other hand, not all eukaryotic tRNAsAla bear a G3:U70 base pair in their acceptor stems. One such example was found in Drosophila melanogaster, where the cytoplasmic tRNAAla has a canonical G3:U70 base pair in its acceptor stem, but the GU base pair has been translocated to position 2:71 in its mitochondrial isoacceptor. As a result, the mitochondrial tRNAAla (with a G2:U71 base pair) can only be recognized by its mitochondrial cognate enzyme (35). Further investigation revealed that the ability to recognize the translocated GU base pair is conferred by an insertion peptide of 27 amino acid residues embedded inside the mitochondrial enzyme (35). Despite V. polyspora also possessing two distinct AlaRS genes, no similar insertion peptide was found in their protein products, and all of their cognate tRNAs contained a G3:U70 base pair (Figure 2).
An earlier phylogeny proposed that the existing AlaRS gene in mitochondrion-containing eukaryotes was horizontally transferred to the nucleus from an ancestral mitochondrion and therefore should be regarded as being of mitochondrial origin (36,37). However, the AlaRS gene in the amitochondriate protists G. lamblia and T. vaginalis appears to have evolved from a different origin. In fact, the AlaRS sequences retrieved from these two eukaryotic organisms were clustered into a sister group that grossly deviated from the major eukaryotic branch and exhibited higher affinity to the archaeal branch instead (36) (Figure 7). Conceivably, the evolutionary history of AlaRS in eukaryotes is far more sophisticated than anticipated. In this regard, our findings reported herein are of particular interest, as they show that all yeast AlaRS genes, including those in V. polyspora, are of mitochondrial origin, regardless of the cellular compartments to which their protein products are targeted (Figure 7). As all other yeast species studied contain a single dual-functional AlaRS gene (Figures 1–3), it is likely that contemporary AlaRS genes in V. polyspora arose from duplication of such a gene. Notably, only one copy of the AlaRS gene was retained in the genomes of S. cerevisiae and C. glabrata after the sorting-out process (Figures 1 and 7). Moreover, it was shown that only a single copy of the valyl-tRNA synthetase gene was retained in yeast species descended from the aforementioned event (11). Thus, the evolutionary footprints in relation to genome duplication and reduction appear to be independent for each aminoacylation activity, even in the same organism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Science Council (Taipei, Taiwan; NSC97-2311-B-008-002-MY3, NSC97-2311-B-008-003-MY3 to C.C.W.); Armed Forces Taoyuan General Hospital (Lungtan, Taiwan; to C.Y.K., C.C.W.). Funding for open access charge: National Science Council (Taiwan).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Burbaum JJ, Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J. Biol. Chem. 1991;266:16965–16968. [PubMed] [Google Scholar]
- 2.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 3.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietrich A, Weil JH, Marechal-Drouard L. Nuclear-encoded transfer RNAs in plant mitochondria. Annu. Rev. Cell Biol. 1992;8:115–131. doi: 10.1146/annurev.cb.08.110192.000555. [DOI] [PubMed] [Google Scholar]
- 5.Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang HL, Yeh LS, Chen NK, Ripmaster T, Schimmel P, Wang CC. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 2004;279:49656–49663. doi: 10.1074/jbc.M408081200. [DOI] [PubMed] [Google Scholar]
- 7.Chang KJ, Wang CC. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:13778–13785. doi: 10.1074/jbc.M311269200. [DOI] [PubMed] [Google Scholar]
- 8.Natsoulis G, Hilger F, Fink GR. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- 9.Chatton B, Walter P, Ebel JP, Lacroute F, Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 1988;263:52–57. [PubMed] [Google Scholar]
- 10.Souciet G, Menand B, Ovesna J, Cosset A, Dietrich A, Wintz H. Characterization of two bifunctional Arabdopsis thaliana genes coding for mitochondrial and cytosolic forms of valyl-tRNA synthetase and threonyl-tRNA synthetase by alternative use of two in-frame AUGs. Eur. J. Biochem. 1999;266:848–854. doi: 10.1046/j.1432-1327.1999.00922.x. [DOI] [PubMed] [Google Scholar]
- 11.Chiu WC, Chang CP, Wen WL, Wang SW, Wang CC. Schizosaccharomyces pombe possesses two paralogous valyl-tRNA synthetase genes of mitochondrial origin. Mol. Biol. Evol. 2010;27:1415–1424. doi: 10.1093/molbev/msq025. [DOI] [PubMed] [Google Scholar]
- 12.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cigan AM, Pabich EK, Donahue TF. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baim SB, Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cigan AM, Donahue TF. Sequence and structural features associated with translational initiator regions in yeast–a review. Gene. 1987;59:1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen SJ, Lin G, Chang KJ, Yeh LS, Wang CC. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J. Biol. Chem. 2008;283:3173–3180. doi: 10.1074/jbc.M706968200. [DOI] [PubMed] [Google Scholar]
- 17.Chang CP, Chen SJ, Lin CH, Wang TL, Wang CC. A single sequence context cannot satisfy all non-AUG initiator codons in yeast. BMC Microbiol. 10:188. doi: 10.1186/1471-2180-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang KJ, Lin G, Men LC, Wang CC. Redundancy of non-AUG initiators. A clever mechanism to enhance the efficiency of translation in yeast. J. Biol. Chem. 2006;281:7775–7783. doi: 10.1074/jbc.M511265200. [DOI] [PubMed] [Google Scholar]
- 19.Abramczyk D, Tchorzewski M, Grankowski N. Non-AUG translation initiation of mRNA encoding acidic ribosomal P2A protein in Candida albicans. Yeast. 2003;20:1045–1052. doi: 10.1002/yea.1020. [DOI] [PubMed] [Google Scholar]
- 20.Diaz-Lazcoz Y, Aude JC, Nitschke P, Chiapello H, Landes-Devauchelle C, Risler JL. Evolution of genes, evolution of species: the case of aminoacyl-tRNA synthetases. Mol. Biol. Evol. 1998;15:1548–1561. doi: 10.1093/oxfordjournals.molbev.a025882. [DOI] [PubMed] [Google Scholar]
- 21.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 22.Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang HY, Kuei Y, Chao HY, Chen SJ, Yeh LS, Wang CC. Cross-species and cross-compartmental aminoacylation of isoaccepting tRNAs by a class II tRNA synthetase. J. Biol. Chem. 2006;281:31430–31439. doi: 10.1074/jbc.M601869200. [DOI] [PubMed] [Google Scholar]
- 24.Wang CC, Chang KJ, Tang HL, Hsieh CJ, Schimmel P. Mitochondrial form of a tRNA synthetase can be made bifunctional by manipulating its leader peptide. Biochemistry. 2003;42:1646–1651. doi: 10.1021/bi025964c. [DOI] [PubMed] [Google Scholar]
- 25.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 26.Ripmaster TL, Shiba K, Schimmel P. Wide cross-species aminoacyl-tRNA synthetase replacement in vivo: yeast cytoplasmic alanine enzyme replaced by human polymyositis serum antigen. Proc. Natl Acad. Sci. USA. 1995;92:4932–4936. doi: 10.1073/pnas.92.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 28.McClain WH, Foss K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a ‘variable pocket’. Science. 1988;241:1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- 29.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 30.Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- 31.Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hann SR, Sloan-Brown K, Spotts GD. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992;6:1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- 33.Packham G, Brimmell M, Cleveland JL. Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem. J. 1997;328(Pt 3):807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schimmel P, Giege R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl Acad. Sci. USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovato MA, Chihade JW, Schimmel P. Translocation within the acceptor helix of a major tRNA identity determinant. EMBO J. 2001;20:4846–4853. doi: 10.1093/emboj/20.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chihade JW, Brown JR, Schimmel PR, Ribas De Pouplana L. Origin of mitochondria in relation to evolutionary history of eukaryotic alanyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2000;97:12153–12157. doi: 10.1073/pnas.220388797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.