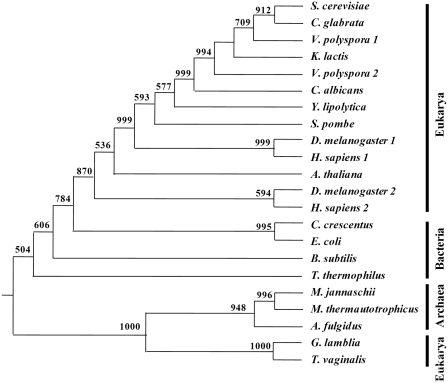
Figure 7.
Phylogenetic analysis of the relationships of VpAlaRS1, VpAlaRS2 and other AlaRSs. Sequences comprising the core active sites of AlaRSs were aligned with Clustal W (38) and analyzed using the Neighbor-joining method. Numbers at the nodes denote bootstrapping frequencies calculated from 1000 trees. A. fulgidus, Archaeoglobus fulgidus (NP_071080); A. thaliana, Arabidopsis thaliana (CAA80380); B. subtilis, Bacillus subtilis (NP_390618); C. crescentus, Caulobacter crescentus (NP_421332); D. melanogaster, Drosophila melanogaster (DmAlaRS1, NP_523511.2; DmAlaRS2, NP_523932.2); E. coli, Escherichia coli (NP_417177); G. lamblia, Giardia lamblia (XP_001704452); H. sapiens, Homo sapiens (HsAlaRS1, BAA06808; HsAlaRS2, NP_065796); M. jannaschii, Methanocaldococcus jannaschii (NP_247543); M. thermautotrophicus, Methanothermobacter thermautotrophicus (NP_276794); T. thermophilus, Thermus thermophilus (YP_145097); T. vaginalis, Trichomonas vaginalis (AAT95880.1).