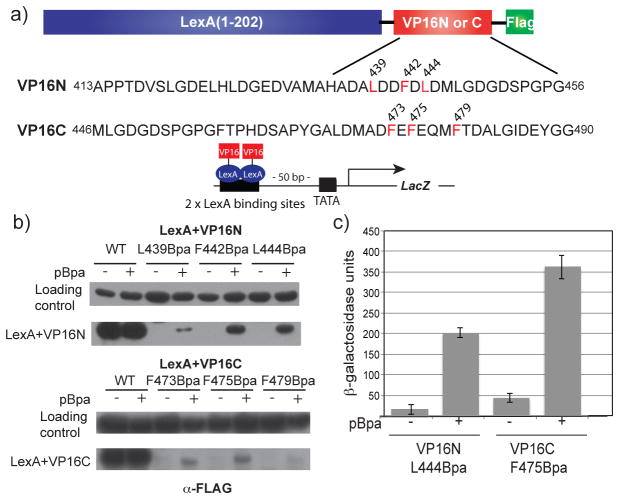
Figure 2.
Incorporation of Bpa within the VP16 TAD. (a) Plasmids encoding the DNA binding domain (DBD) of LexA fused to either the N- or C-terminal VP16 TAD as well as a FLAG tag were constructed. The LexA DBD was utilized to exclusively examine transcriptional activation at the 2 unique LexA binding sites upstream of the LacZ reporter in S. cerevisiae. Positions at which Bpa mutagenesis was carried out are within regions of the VP16N or VP16C subdomains known to participate in coactivator binding (sites of incorporation highlighted in red). The loading control is an approximately 71 kDa, FLAG-detected yeast protein whose expression level does not vary with activator identity.(22) (b) Yeast cells bearing plasmids encoding the various LexA+VP16 constructs and the Bpa specific tRNA/synthetase pair expressed by pSNRtRNA-pBpaRS were grown in the presence or absence of 1 mM Bpa and analyzed by Western blot (c) LexA+VP16N L444Bpa and LexA+VP16C F475Bpa were assessed for their ability to upregulate transcription of an integrated LacZ reporter gene in S. cerevisiae as measured by liquid β-galactosidase assays. Each activity is the average of values from at least three independent experiments with the indicated error (SDOM). See Supporting Information Figure S7 for activity assays of the remaining mutants.