Figure 3. Identification of AMPK phosphorylation sites in four AMPKα2 substrates.
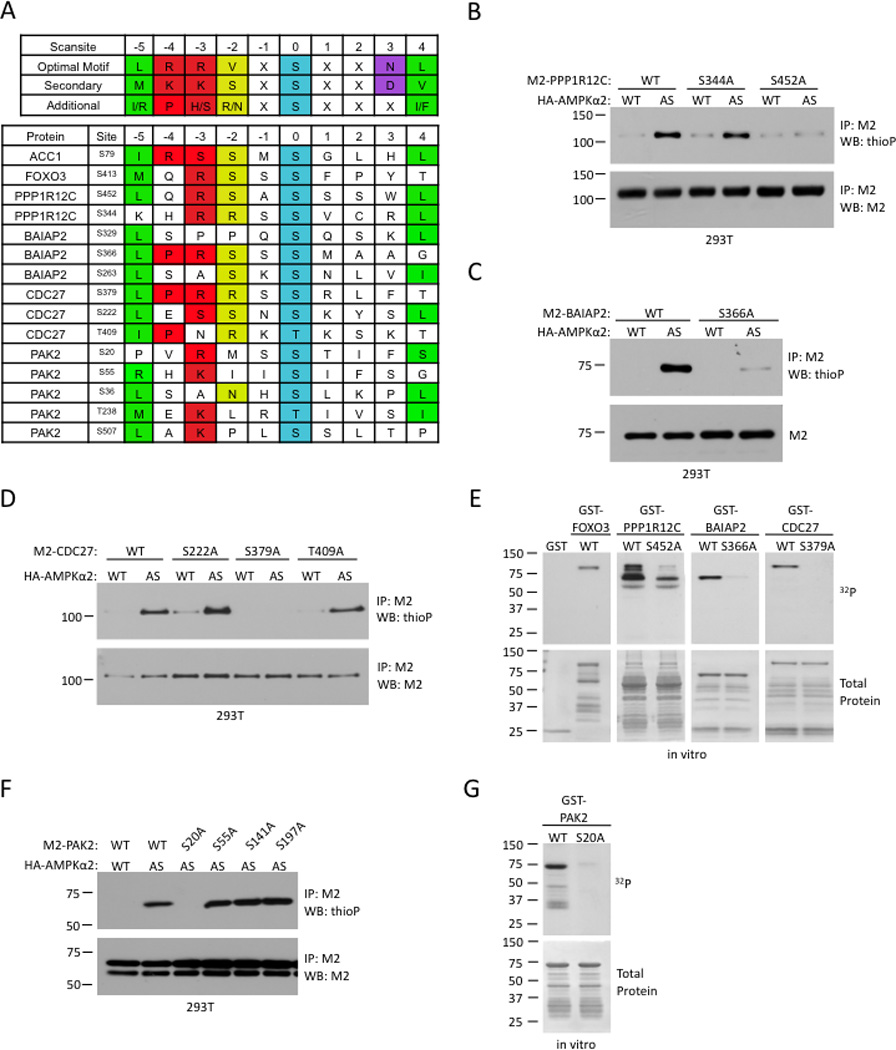
(A) Scansite prediction of potential AMPK phosphorylation sites. The AMPK consensus phosphorylation motif, derived from Gwinn et al (2008), is shown on the top in single letter amino acid code. Positions denoted as ‘X’ showed some additional modest selectivities among amino acids, but lack the strong discrimination shown in the specified positions. Known AMPK phosphorylation sites in ACC1 and FOXO3 are shown as comparisons.
(B)–(D) In vivo identification of AMPKα2 phosphorylation sites in PPP1R12C (B), BAIAP2 (C), and CDC27 (D). Analysis was carried out as in Figure 1D.
(E) In vitro validation of AMPK phosphorylation sites in PPP1R12C, BAIAP2, and CDC27. Kinase assays were carried out as in Figure 2E.
(F) In vivo testing of putative phosphorylation sites identified by tandem mass spectrometry for PAK2. Analysis was carried out as described in Figure 1D.
(G) In vitro validation of AMPK phosphorylation sites in PAK2. Kinase assay was carried out as in Figure 2E. All panels are representative of 2 independent experiments.
